A team of scientists from the USA recently developed a recombinant “Angiotensin converting enzyme 2 (ACE2) Triple Decoy” that can trap severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and prevent its propagation inside the host cell. They have reported that the decoy has high affinity for different mutants of the spike receptor-binding domain (RBD) known to significantly increase viral transmission. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Since its emergence in December 2019, SARS-CoV-2 has undergone more than 12000 mutations that collectively lead to the emergence of various genetic variants of the original virus. According to the available literature, some of these viral variants are more transmissible and may be more lethal than the original SARS-CoV-2 strain. These variants are collectively termed as the Variants of Concern (VOC) because of their ability to escape host humoral immune responses developed either by natural SARS-CoV-2 infection, or by therapeutic monoclonal antibodies and vaccines. Because the majority of anti-SARS-CoV-2 antibodies preferentially target the spike RBD, mutations emerging in this domain could potentially affect the binding affinity and neutralizing efficacy of these antibodies. Moreover, there is evidence suggesting that antibodies developed via natural infection or vaccination can actually trigger that process of viral evolution, leading to the emergence of new viral variants with improved fitness against the host immune responses.
Given the significant impact of viral mutations on antibody-driven therapeutic approaches, the scientists of the current study have developed an ACE2 decoy that is less likely to be affected by viral mutations. The decoy would compete with endogenous ACE2 for binding to spike RBD, leading to the entrapment of the virus and inhibition of viral replication.
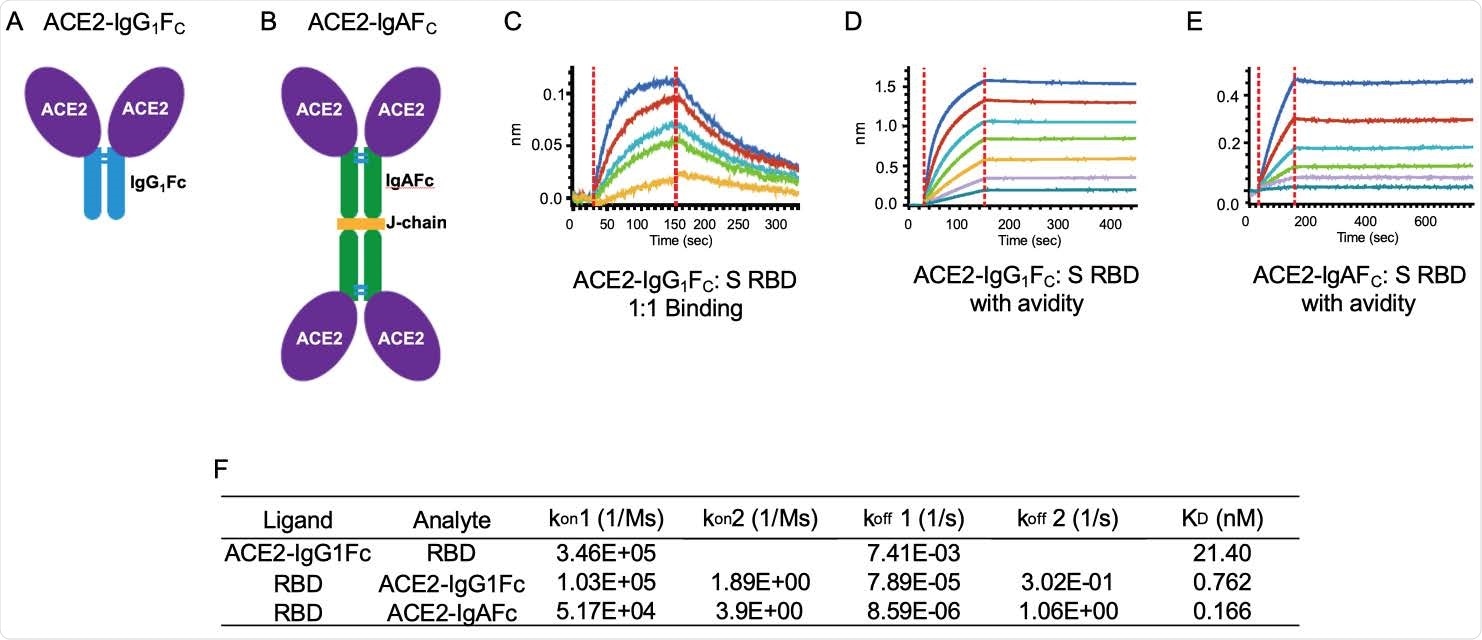
ACE2-IgG1FC and dimeric -IgAFC decoys bind the spike receptor-binding domain (S RBD) with high affinity. The (A) ACE2-IgG1FC decoy; (B) dimeric ACE2-IgAFC decoy fused via a chain are shown. Biolayer Interferometry (BLI) kinetics analysis of (C) 1:1 binding and (D) binding with avidity for the ACE2-IgG1FC decoy; and (E) BLI binding with avidity for the ACE2-IgAFC decoy are shown. (F) Table of binding affinity values.
Study design
The scientists conducted molecular dynamic simulation studies to predict potential ACE2 mutations that significantly increase the spike RBD binding affinity of ACE2. For a successful competitive binding, an ACE2 decoy should have higher affinity than endogenous ACE2. By analyzing these mutations, they developed and screened several recombinant ACE2-IgG1FC or -IgAFC fusion proteins as potential decoy candidates. Their screening led to the identification of an ACE2 decoy with two mutations (T27Y/H34A), which showed robust spike RBD binding affinity and SARS-CoV-2 neutralizing ability.
To inhibit the enzymatic activity of ACE2, they further modified the decoy by adding another mutation (H374N). Finally, they tested the efficacy of the ACE2 triple decoy (T27Y/H34A/H374N) against spike RBD variants expressing E484K, K417N, N501Y, and L452R mutations.
Important observations
By conducting binding assays, the scientists observed that the combination of T27Y and H34A mutations increased the spike RBD binding affinity of the decoy by 35-fold. Similarly, by conducting a live SARS-CoV-2 neutralization assay, they observed that the decoy containing these two mutations had a 15-fold higher virus-neutralizing ability than the wildtype decoy without any mutation. For a mechanistic evaluation, they analyzed the simulation data and noticed that a threonine (T) to tyrosine (Y) substitution at residue 27 of ACE2 caused favorable hydrophobic interactions with the RBD. Similarly, a histidine (H) to alanine (A) substitution at residue 34 increased the surface area for RBD – ACE2 interaction.
Because administration of an enzymatically active ACE2 decoy can cause severe adversities at the physiological level, they introduced the H374N mutation to the double-mutant decoy and observed that the mutation significantly inhibited the ACE2 enzymatic activity without hampering its spike RBD binding affinity.
To examine the binding affinity of ACE2 triple decoy, they selected a panel of RBD mutations present in currently circulating VOCs. Their analysis revealed that the binding affinity of triple decoy increased significantly for the spike RBD expressing N501Y or L452R mutation. However, the highest affinity was observed for the spike variant expressing both E484K and N501Y mutations.
Furthermore, they conducted a surrogate SARS-CoV-2 neutralization assay to examine whether the triple decoy is able to inhibit the interaction of wildtype and mutated RBDs with wildtype ACE2. The findings revealed that the triple decoy is able to compete with wildtype ACE2 in the presence of both wildtype and mutated spike RBD variants. For the final confirmation, they conducted additional simulation studies that revealed a strong affinity of the ACE2 decoy for both wildtype and mutated spike RBD.
Study significance
The study describes the development process and evaluation of an ACE2 decoy that harbors certain ACE2 mutations for an improved spike RBD binding affinity. Given its strong binding affinity for several spike RBD variants, the decoy can be potentially used as a therapeutic as well as a prophylactic to control SARS-CoV-2 transmission.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources