An interesting new study deals with the effects of the natural mutations in the spike antigen of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen that is responsible for the ongoing coronavirus disease pandemic.
The inability to control the spread of this virus has been linked to the rapid emergence of new mutations that materially affect its transmissibility, infectivity, and virulence. This phenomenon also threatens the goal of achieving population immunity via vaccination-induced antibodies.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The current study, available in preprint form on the bioRxiv* server, deals with four SARS-CoV-2 variants with unique sets of mutations, exploring their impact on biological activity.
Virus-host interactions
The virus engages the host cell receptor, the angiotensin-converting enzyme 2 (ACE2), via its spike glycoprotein, a protein that protrudes from its membrane. This is composed of an S1 and S2 subunit, with a novel furin cleavage site at the interface.
The S1 is responsible for spike-ACE2 binding. It contains the N-terminal domain (NTD), the ACE2 receptor-binding domain (RBD), and two subdomains (SD1 and SD2). The NTD and the RBD are dominant antigens for neutralizing antibodies.
The RBD is metastable, with an ‘up’ and a ‘down’ conformation. Only the latter is accessible to ACE2 binding.
The S2 has a second cleavage site, the S2’ site, which is attacked by the serine protease TMPRSS2. It also contains a fusion peptide (FP), heptad repeat 1 (HR1), the central helix (CH), the connector domain (CD), heptad repeat 2 (HR2), the transmembrane domain (TM) and a cytoplasmic tail (CT).
Spike-ACE2 engagement is followed by protease-mediated cleavage at the S1/S2 interface and the S2 site. This leads to large conformational changes in the spike, causing membrane-cell fusion and viral entry into the host cell.
Late in 2020, several new variants of concern (VOCs) appeared, which were resistant to neutralizing antibodies and more infectious than earlier variants. These carried the dominant D614G mutation in the spike but also other sets of novel mutations. The four variants studied here are the mink variant, the UK variant, the B.1.1.28 variant and the South African variant.
Mink-associated SARS-CoV-2 variant
The outbreak of SARS-CoV-2 infection among mink farmworkers in early 2020 was followed by the occurrence of similar strains in many other countries, leading eventually to the culling of all 17 million minks farmed in Denmark.
The variant has five spike mutations, H69 (H69Δ) and V70 (V70Δ) deletion in the NTD, and Y453F in the RBD, I692V downstream of the furin cleavage site in SD2, and M1229I in the transmembrane domain. The last was not included, however, in the virus construct.
This variant showed a 3.5-fold higher binding affinity to ACE2 compared to the original D614G ectodomain. This was due mainly to a lower rate of dissociation of the bound spike-ACE2 complex, caused by the Y453F mutation.
The variant bound strongly to RBD-specific neutralizing antibodies, DH1041, DH1043 and DH1047, and increased binding affinity with the neutralizing NTD-targeting antibodies DH1050.1 and DH1050.2 by 3.5 and 2.6-fold, respectively. This was mainly due to an increased on-rate, and was contributed chiefly by the H69/V70Δ mutation.
Despite the increase in receptor binding affinity, immune evasion was not observed.
The researchers also found that the 3 RBD-down states were unexpectedly variable, and the S2 subunit that was known to remain invariant in previously reported prefusion spike structures showed conformational changes. Their examination also showed a new state that lacked the S1 subunit of one of the three protomers.
These findings, put together, indicate that the prefusion spike structure might be destabilized by the mutations in this variant.
The B.1.1.7 variant
This variant, termed the UK variant, is thought to be more transmissible, more virulent and more dangerous in terms of mortality than the D614G variant. It contains eight spike mutations, including N501Y in the receptor-binding motif in the RBD, ΔH69/V70 and ΔY144 deletions in the NTD, A570D in SD1, P681H proximal to the furin cleavage site on SD2, and three others.
This variant showed a five-fold increase in ACE2 binding, due mainly to the N501Y mutation. “This is consistent with reports identifying residue 501 as a hotspot for mutagenic modulation of ACE2 binding affinity.”
This variant is susceptible to vaccine-elicited antibodies and to the RBD-targeting natural antibodies above. Conversely, it is resistant to NTD-directed antibodies such as 4A8, 5-24, 4-8 and DH1050.1, an effect linked to the NTD mutation ΔY144.
Structural studies showed the allosteric effect of the paired mutations, the S982A [in the HR1 region] and A570D in the spike regions distal to the RBD and NTD. The various changes in the spike structure result in disengagement of the NTD from the adjacent RBD, which accounts for the increased RBD mobility and exposure.
This indicates pairing acts as an allosteric switch through coupling of domain movements.”
The B.1.351 and B.1.1.28 variants
The B.1.351 variant has eight mutations in the spike protein, seven of which affect the NTD and RBD. K417N, E484K and N501Y are in the RBD. In addition to these three, the B.1.1.28 variant has other mutations. In particular, the P.1 variant arose from this lineage and has caused a fresh wave of cases in an area of Brazil called Manaus, where 75% of the people had already been infected.
When mutations accumulate in these regions, encoding the dominant antigens eliciting the immune response, they are likely to affect positive selection pressure.
Both these variants show improved ACE2 binding affinity because of the N501Y mutation. The E484K and the triple RBD mutation in the B.1.1.28 variant both failed to affect binding to the neutralizing NTD-directed antibody DH1050.1. However, it did affect binding in a construct with multiple NTD mutations.
Binding to RBD-targeting antibodies DH1041 and DH1043 was robust but moderately decreased with B.1.1.28, even though the E484K mutation is at the epitope or binding interface of these antibodies. Conversely, it was markedly reduced for B.1.351, indicating that the NTD mutations produce allosteric inhibition of antibody binding.
The B.1.351 variant was observed to be resistant to some antibodies targeting the RBD and NTD, and to convalescent sera as well. This is due to the three RBD mutations, especially the E484K mutation. This reduces or abolishes binding to a number of class I antibodies targeting the RBD, such as DH1041.
E484K alters RBD conformation to produce immune evasion
Cryo-EM (cryo-electron microscopy) showed that the three mutations in the RBD cause disorder in the RBDs in the 3-RBD-down state, thus favoring the ‘up’ state. The E484K mutation leads to increased disorder in the RBD hook, which may also cause the increased RBD ‘up’ propensity by weakening the coupling between adjacent RBDs.
Both the RBD-RBD interactions in the ‘up’ state, as well as the bound antibody-adjacent RBD contacts, are likely to be implicated in stabilizing the antibody binding to mutants with E484K.
The B.1.351 and B.1.1.28 variants may utilize modification of the previously identified N343-glycan gate that contacts the adjacent RBD tip at a location near the E484K mutation to achieve [enhanced RBD exposure].”
The main outcome of changes in the 3-RBD-down state, from this series of structural studies, appears to be the increased exposure of the RBD, primarily by destabilizing one or more of the RBDs.
Different mechanisms converge to enhance the exposure of the RBD. With the B.1.351 and B.1.1.28 variants, the destabilization is due to inter-protomer contacts. With the other two, it is mediated by modifications in the interactions between S1/S2 with the S2 of an adjacent RBD.
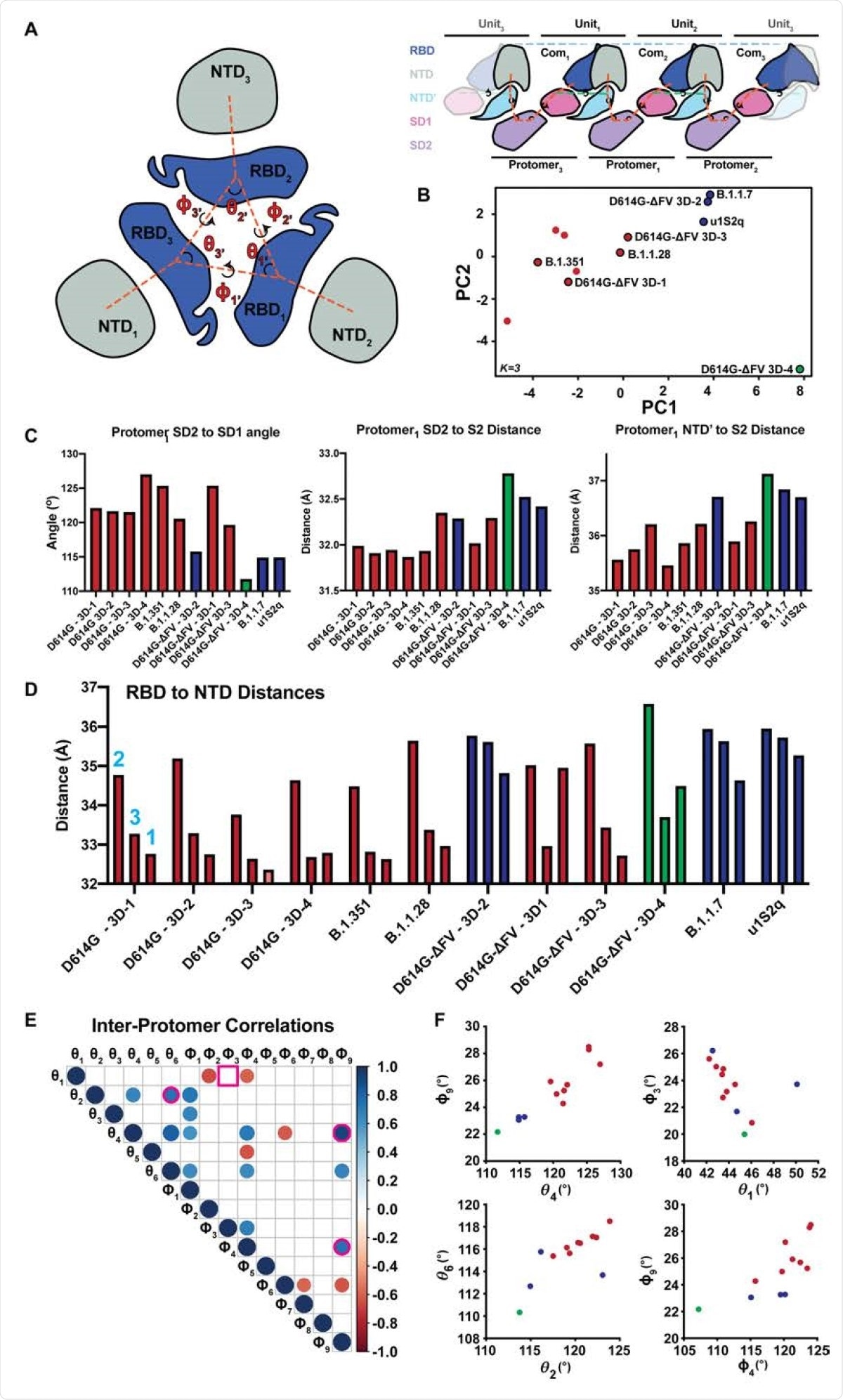
Comparison of inter-protomer network and RBD to RBD vector measures. A. (left) RBD and NTD vectors, angles, and dihedrals. (right) Simplified schematic of the SD2, SD1, NTD' inter-protomer contact network. B. Principal components analysis of the inter-protomer network and RBD to RBD vector measures colored according to k-centers (K=3). Clusters correspond to a GSAS-D614G (D614G) like cluster (red), a u1S2q like cluster (blue), and outlier S-GSAS-D614G-ΔFV (D614G-ΔFV) 3D-4 (green). C. Top three contributors to PCA component one. D. RBD to NTD distance for the variants including D614G-ΔFV and the previously determined D614G. E. Significant Correlations between the inter-protomer angle measures (N=12, p<0.05). Pink outlines identify relationships plotted in panel (F). Square outline identifies non-significant correlation in the full structure set that was significant in the D614G cluster only correlations. F. Selected vector relationship plots. Colors are according to clusters in the PCA analysis in panel (B).
What are the implications?
All the new variants showed increased binding affinity to the ACE2 receptor. However, the changes that occurred in the SARS-CoV-2 spike within humans are different from those that emerged during mink-human transmission.
In humans, the increased ACE2 binding caused increased transmissibility. In the mink-associated variant, the increased binding affinity may overcome the natural decrease in affinity for the homologous receptor in minks, thus helping the virus to adapt to the mink host.
This variant may have lost fitness in this process, as it retained the original susceptibility to antibody neutralization. The postulated loss of fitness is supported by the instability of the spike in this variant, including a population of partly unfolded spikes. These findings may also explain how the mink variant failed to spread widely among humans.
The variants that emerged during spread among humans only showed different mechanisms of spike adjustment in the NTD and RBD, producing a convergent destabilization of the 3-RBD-down state. Since the functional conformation of the RBD in coronaviruses depends on a network of S1 subunit domain interactions, and since the spike protein is metastable, small changes in the domain pairing strength produce large changes in the configuration of the whole molecule.
This mechanism is seen to be at work in the emerging VOCs, disrupting the spike structural state. Mutations in the spike protein may affect RBD conformation via allosteric effects, which in turn is influenced by the subdomains.
This structural study thus provides insights into how these natural mutations affected the spike conformation and viral biology.
We demonstrate that convergent S protein evolution to increase SARS-CoV-2 transmissibility and escape neutralization at immunodominant NTD and RBD epitopes can occur via different allosteric communication networks in the spike.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gobeil, S. et al. (2021). Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. bioRxiv preprint. doi: https://doi.org/10.1101/2021.03.11.435037, https://www.biorxiv.org/content/10.1101/2021.03.11.435037v1
- Peer reviewed and published scientific report.
Gobeil, Sophie M.-C., Katarzyna Janowska, Shana McDowell, Katayoun Mansouri, Robert Parks, Victoria Stalls, Megan F. Kopp, et al. 2021. “Effect of Natural Mutations of SARS-CoV-2 on Spike Structure, Conformation, and Antigenicity.” Science 373 (6555). https://doi.org/10.1126/science.abi6226. https://www.science.org/doi/10.1126/science.abi6226.