Betacoronavirus (β-CoVs) are positive-stranded RNA viruses having zoonotic potential. This enveloped virus infects a wide range of hosts that includes humans. Typically, these viruses infect the respiratory tract, and the infection varies from mild to severe with flu-like symptoms. Researchers revealed that among CoVs, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the most infectious strain, and the causal agent of the COVID-19 pandemic.
CoVs are enveloped by a protein membrane that consists of a minimum of three structural proteins, namely, Spike (S), Membrane (M), and Envelope (E). They also contain accessory factors such as Orf3a. E and M proteins are essential for viral assembly, whereas, S protein mediates the attachment of the virus to the host cell receptors, and also takes part in the transfer of the viral genome to the host. The primary function of the S protein is infectivity. Because of this it is the target site of most vaccines.
The synthesis of E, M, and S proteins occurs in the ribosomes associated with the endoplasmic reticulum (ER). Folding and quaternary assembly (trimerization of Spike) occur in these cell organelles. The post-translation modifications, e.g., N-glycosylation, also occur on their luminal domains and S-acylation on the cytosolic domains.
Scientists have reported that in viruses such as SARS-CoV-1 and mouse hepatitis virus (MHV), both S and E proteins undergo S-acylation. S-acylation is regarded as a hallmark of viral envelope proteins. It chemically links, via a covalent bond, a protein to the sulfur atom of cytosolic cysteines.
Even though extensive research is being carried out across the globe to study different aspects of the virus and find out means to contain the infection, there is a gap in research regarding the accurate molecular understanding of the biogenesis of the virus and the factors responsible for the infectivity.
A new study published on the bioRxiv* server focuses on the role of S-acylation in the SARS-CoV-2 infection cycle. It has also characterized Spike acylation and has also identified other acylated viral proteins and host enzymes involved in the process of infection. Further, it has also revealed the importance of acylation in viral biogenesis and infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In this study, researchers have used two assays to monitor protein acylation. The first assay is the Acyl-Resin Assisted Capture assay (Acylrac). The second assay involves incubating cells with radioactive 3H-palmitate, thereby examining the incorporation by Spike using immunoprecipitation and autoradiographic analysis. In order to determine the acyltransferases responsible for the modification of Spike, researchers have individually depleted the 23 conserved zinc-finger domains with a DHHC motif in their catalytic sites (ZDHHCs) by RNA interference in Hela cells. Thereby, the researchers have identified ZDHHC20 as the key acyltransferase responsible for the lipidation of SARS-CoV-2 Spike, E, and Orf3a.
SARS-CoV-2 Spike consists of ten conserved cysteine residues within its first 20 cytosolic amino acids. While studying the importance of these cysteine residues, researchers found that these are targets of S-acylation and lipidation of those closest to the membrane required to bring the distal cysteines closer to the membrane, making it more accessible to the ZDHHC active site. Further, this research has also shown that following synthesis, acylation of the S protein takes place in the ER by ZDHHC20.
Researchers have also studied how S-acylation modifies the turnover rate of proteins. The first method involves 35S Cys/Met metabolic pulse-chase experiments, where they found 35S Cys/Met metabolic labeling time was short (20 min). Additionally, they found a rapid degradation of spike protein following its synthesis when ZDHHC enzymes were silenced. Thereby, this research shows acylation is vital for efficient Spike biogenesis.
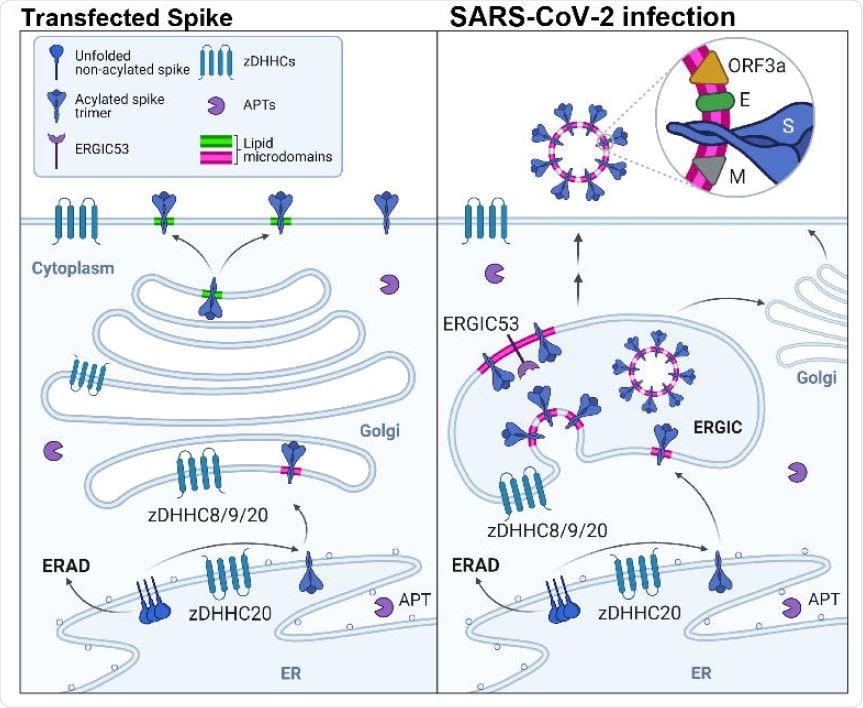
This research has highlighted some of the vital contributions of Spike acylation. These include a) induction of biogenesis by protecting it from premature targeting to ER degradation pathway, b) an increase in Spike half-life, and c) regulating the lipid composition of its immediate membrane environment.
Researchers have conducted lipidomic analyses which reveal the unique lipid composition of SARS-CoV-2 virions. The analyses have revealed the presence of a high concentration of specific early Golgi sphingolipids, which is a unique feature that is absent in other viruses budding in the plasma membrane. One of the most important outcomes of this research is the revelation of the importance of S-acylation and sphingolipid metabolism in promoting the SARS-CoV-2 infection.
To conclude, this research highlights that S-acylation and the ZDHHC20 enzyme can be potential drug targets for the treatment of coronavirus infection. Previous research has also indicated that ZDHHC20 is associated with acylation of the hemagglutinin of the Influenza virus. Further analysis of its structure also indicates that this acyltransferase can be a possible target for anti-viral drugs. This research also points out that S-acylation and lipid biogenesis-related enzymes can be novel potential anti-viral targets.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
S-acylation controls SARS-Cov-2 membrane lipid organization and enhances infectivity, Francisco. Sarmento Mesquita, Laurence Abrami, Oksana Sergeeva, Priscilla TurelliBeatrice Kunz, Charlene Raclot, Jonathan Paz Montoya, Luciano Abriata, Matteo Dal Peraro, Didier Trono, Giovanni D'Angelo, F. Gisou van der Goot, bioRxiv 2021.03.14.435299; doi: https://doi.org/10.1101/2021.03.14.435299, https://www.biorxiv.org/content/10.1101/2021.03.14.435299v1
- Peer reviewed and published scientific report.
Mesquita, Francisco S., Laurence Abrami, Oksana Sergeeva, Priscilla Turelli, Enya Qing, Béatrice Kunz, Charlène Raclot, et al. 2021. “S-Acylation Controls SARS-CoV-2 Membrane Lipid Organization and Enhances Infectivity.” Developmental Cell 56 (20): 2790-2807.e8. https://doi.org/10.1016/j.devcel.2021.09.016. https://www.cell.com/developmental-cell/fulltext/S1534-5807(21)00734-6.