The coronavirus disease (COVID-19) pandemic has sparked a worldwide crisis since its emergence in late December 2019 in Wuhan, China. Caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pandemic has taken a toll on the world economy, with millions of people losing their jobs.
In the future, pandemics may cause further severe economic and social burdens. Hence, predicting potential hosts for viruses is crucial.
Researchers at the Department of Biomedical Engineering, College of Engineering, and Center for Quantitative Biology, Peking University, developed a deep learning method that extracts viral genomic features automatically to predict hosts for new viruses, including coronaviruses.
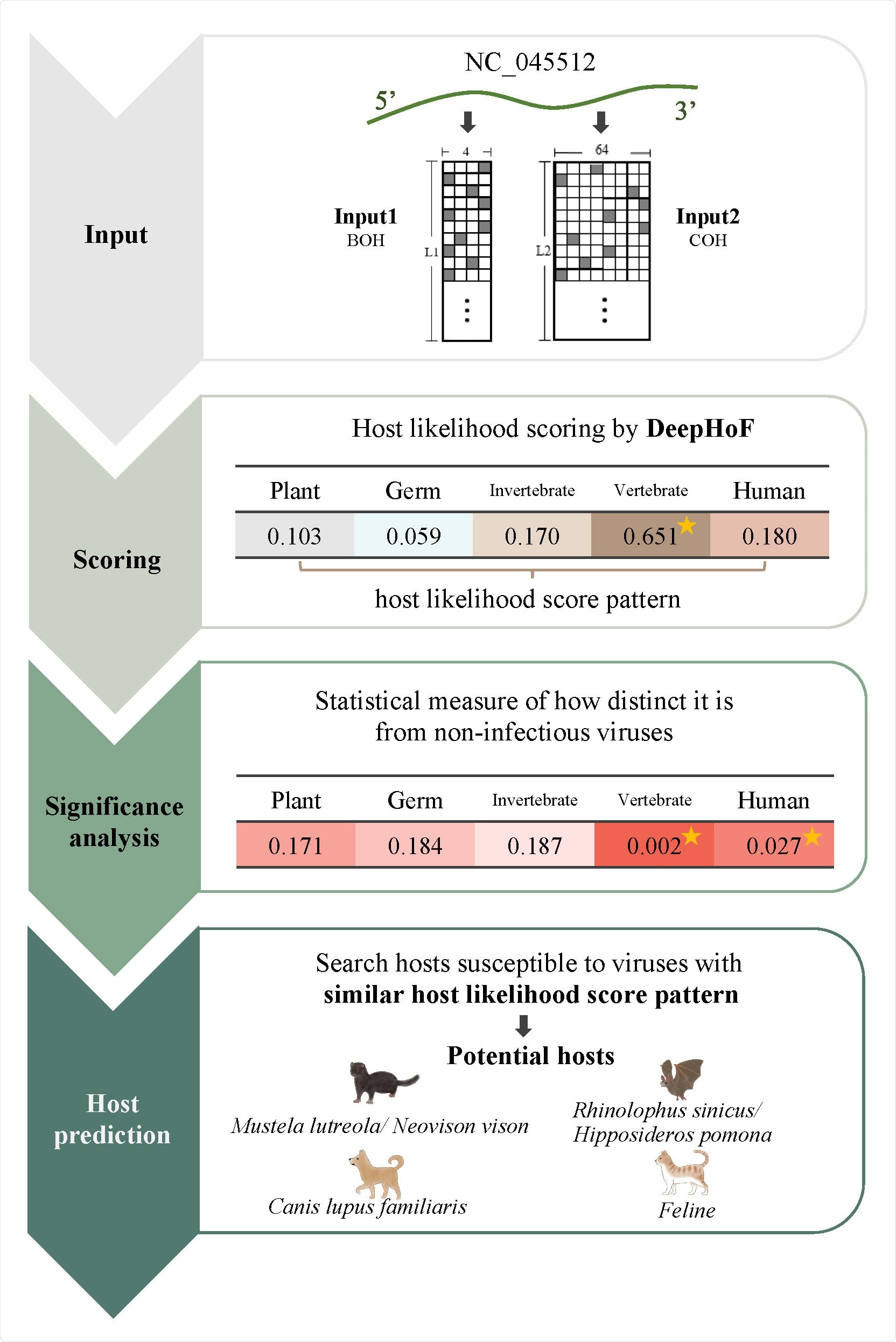
Called DeepHoF (Deep learning-based Host Finder), the deep learning method can predict host likelihood scores on five host types, including germ, plant, invertebrate, non-human vertebrate, and humans.
The study, which appeared on the pre-print server bioRxiv*, revealed how DeepHoF could help prevent future outbreaks that may become as large as the current pandemic.
.jpg)
Study: Predicting Hosts Based on Early SARS-CoV-2 Samples and Analyzing Later World-wide Pandemic in 2020

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study background
When the pandemic first emerged in December 2019, scientists were left baffled on which animal served as the virus's intermediate host. Though monkeys were known to be reservoirs of coronaviruses, determining potential animal carriers can help prevent future outbreaks.
So far, there have been many suggestions for potential hosts of SARS-CoV-2, including pangolins. Other animal cases of SARS-CoV-2 infection were also reported, including dogs, tigers, lions, minks, and cats.
Scientists believe one problem in predicting potential animal hosts of novel viruses is how to implement and improve the capability of computational methods.
New computational methods are based on the similarity of the viral genome composition or host receptor. Hence, they can detect the potential host and pathogenicity of novel viruses.
The study
In the study, the researchers proposed the host prediction algorithm DeepHoF. The method was developed based on BiPath Convolutional Neural Network (BiPathCNN) and can automatically extract the genomic features from the input viral sequences.
The DeepHoF tool was based on extracting viral genomic features automatically to predict host likelihood scores. This can help determine which animals are more likely to become potential viral hosts. DeepHoF made up for the lack of a precise tool applicable to any novel virus, overcoming the limitation of the sequence similarity-based methods.
Visualization of the host likelihood score profiles of SARS-CoV-2 isolates from different GISAID clades and the manually mutated SARS-CoV-2 isolates on two-dimensional PCA SARS-CoV-2 isolates fall into several clear fusiform clusters with different colors according to their clades. Manually mutated with specific marker variants, the 17 earliest sequenced isolates move to the corresponding fusiform cluster of the clade that is represented by the specific marker variants.
The team also performed a deep analysis of the host likelihood profile calculated by DeepHoF. They used the earliest samples of SARS-CoV-2 isolates that give important data in the early viral outbreak.
The researchers inferred that minks, dogs, bats, and cats were potential hosts of SARS-CoV-2, while minks may be one of the most significant hosts. However, the team noted that due to mutations, the isolates' host likelihood score profiles in the long period of the pandemic had slightly changed.
The study findings also revealed a strong link between SARS-CoV-2 isolates collected from the two populations, showing the contribution of mink on higher divergence in SARS-CoV-2. The method's computation showed the uniformity of host range among samples and a strong link of SARS-CoV-2 with humans and minks.
The new method can also determine host ranges for other novel viruses. But there is a limitation in the use of DeepHoF as it does not consider host sequence data, which can be improved in future endeavors.
"Meanwhile, the present study is expected to be further confirmed with both the ongoing events of pandemic and additional experimental findings, and the interpretation of our analysis should be still kept a certain caution," the researchers noted in the study.
The new method can help predict the potential hosts of viruses that may trigger another outbreak or pandemic.
Newly emerged infectious viruses keep threatening the health of various populations. Using computational methods to find pathogenic viruses and knowing the host range can allow for a timely response to prevent future pandemics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Predicting hosts based on early SARS-CoV-2 samples and analyzing later worldwide pandemic in 2020, Qian Guo, Mo Li, Chunhui Wang, Jinyuan Guo, Xiaoqing Jiang, Jie Tan, Shufang Wu, Peihong Wang, Tingting Xiao, Man Zhou, Zhencheng Fang, Yonghong Xiao, Huaiqiu Zhu, bioRxiv, 2021.03.21.436312; doi: https://doi.org/10.1101/2021.03.21.436312, https://www.biorxiv.org/content/10.1101/2021.03.21.436312v1
- Peer reviewed and published scientific report.
Guo, Qian, Mo Li, Chunhui Wang, Jinyuan Guo, Xiaoqing Jiang, Jie Tan, Shufang Wu, et al. 2021. “Predicting Hosts Based on Early SARS-CoV-2 Samples and Analyzing the 2020 Pandemic.” Scientific Reports 11 (1). https://doi.org/10.1038/s41598-021-96903-6. https://www.nature.com/articles/s41598-021-96903-6.