The authors of the study note their epitope antibody test could be helpful and more effective than conventional screening methods to identify people with the most effective plasma and antibodies for antibody therapy in the absence of a vaccine.
How they did it
The team created their antibody test by identifying severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens influencing antibody responses. They expressed the spike protein (S), nucleocapsid protein (N), and the non-structural proteins from the ORF1ab gene. Previous studies suggested they would be the major antigens for humoral immune response. This created 16 coronavirus proteins and 1 human immunoglobulin G (IgG) for positive control.
Spike and nucleocapsid proteins are the main antigenic proteins for SARS-CoV-2
The resulting proteome array was probed with 146 plasma samples acquired from 89 patients that tested positive or negative for SARS-CoV-2. These patients were divided into two groups based on whether they had severe or moderate coronavirus illness. They were further divided into the ‘alive’ or ‘fatal’ group based on whether or not they survived COVID-19 disease.
Groups of patients with moderate or severe illness showed IgG responses to both SARS-CoV-2 spike protein and the nucleocapsid protein, suggesting they are the main antigens in SARS-CoV-2. Patients hospitalized in the ICU had IgG responses specific to spike and nucleocapsid proteins, which increased from day 1 to day 7 and 10 in both the alive and fatal groups. This suggests humoral responses towards the S or N protein tend to become longer-lasting with time.
The fatal group was more likely to exhibit S- or N-specific antibodies. Based on the findings and lack of correlation between the strength of IgG response to S or N and disease severity, the researchers suggest the antibody responses targeting these proteins will not be as accurate in predicting severity or outcome from COVID-19 infection.
Identifying epitopes of interest to SARS-CoV-2 antibody response
The researchers then searched for the exact targets of SARS-CoV-2 antibody responses. They created peptides with 89 epitopes for the S, N and membrane (M) proteins and exposed them to 15 donated plasma samples, including 14 samples from positive infection and one control.
The team found antibody responses honed in on 54 epitopes from the S and N protein. To assess the strength of an antibody response to treatment, the researchers created a ‘master epitope array’ with 16 epitopes that corresponded with strong IgG signaling.
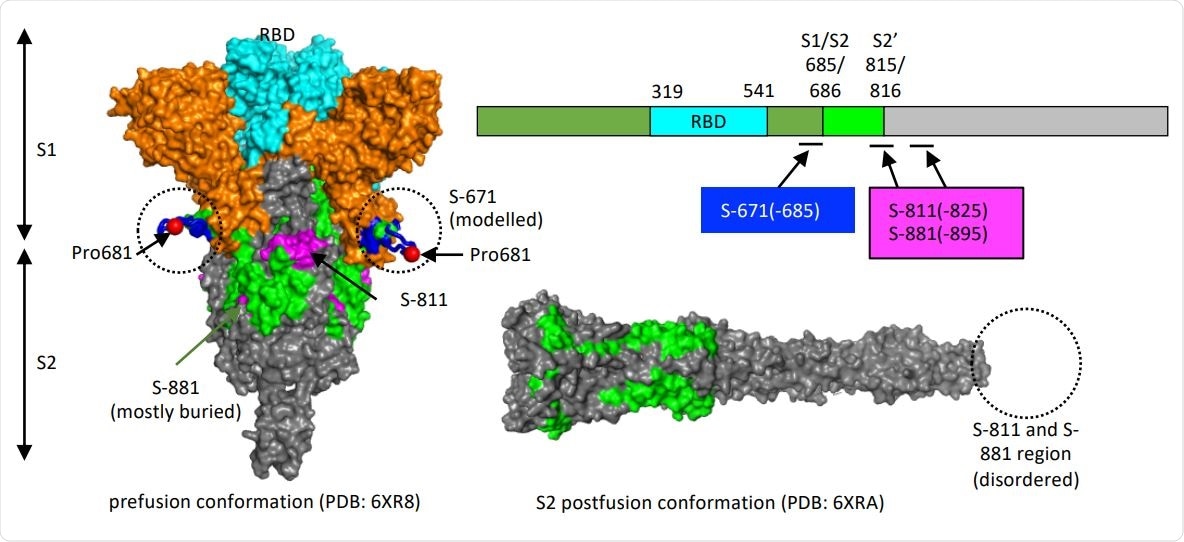
Structure models to show the location of the critical epitopes on the Spike protein. The epitopes S-671, S-811 and S-881 are shown on the domain structure diagram of Spike as well as its prefusion (left) and postfusion (right) conformation. The S protein has two cleavage sites, S1/S2 and S2’. The S-671 epitope is located at the C-terminus of S1, and it is disordered in the prefusion cryo-EM structure (left panel: PDB 6XR8). A homology model from the SWISS-MODEL repository was employed to draw an S671 epitope model in the left panel (colored blue), without cleavage at S1/S2. The Pro681 site is shown with a red sphere. The S2’ cleavage site is located on the S-811 epitope. The S-881 epitope is buried and inaccessible in the prefusion state. However, the region (fusion peptide or FP) targets the host cell membrane and is fully disordered in the post-fusion conformation (right panel: PDB 6XRA, residues 771-911 are disordered). The S1 region is colored orange, except for RBD in cyan. The region between S1/S2 cleavage and S2’ cleavage sites is in green. The S-811 and S-881 epitopes are colored magenta in the prefusion conformation.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The major 16 epitopes were screened with donated plasma from 89 COVID-19 patients and 9 controls. Findings showed that antibodies from people in the severe group in the ICU responded to more epitopes than the group with moderate infection.
The intensity of IgG binding was associated with disease severity and outcome. Moderate illness showed more robust antibody responses to N-156, while long-lasting antibody responses towards S-811 and S-881 were observed in severe infections.
The researchers suggest developing S-811 or S-881 epitopes with severe infection and death could occur because the coronavirus was actively invading cells.
Mutations in epitopes
The coronavirus spike protein has undergone the most mutations out of the whole coronavirus, and is the reason for the emergence of variants. To further understand this, the team created 28 mutations to the S or N proteins, which relates to the B.1.1.7 and B.1351 variant.
Results showed the mutations helped reduce or eliminate IgG binding to epitopes. Specifically, a P681H mutation changed the S-671 epitope and made it undetectable by antibodies.
The team developed a test that would measure the epitope-antibody response for coronavirus infections. When tested against donated plasma, agglutination was only found in plasma that had a positive diagnosis of COVID-19.
Having an S-RBD-dependent antibody response was strongly linked with a favorable outcome. Meanwhile, the test found the S-811—a specific antibody response — was negatively associated with neutralizing power in patients who recovered from infection. But S-811 was positively correlated with severity and fatality. The S-RBD-dependent agglutination efficiency was positively linked with neutralization efficacy in the alive group.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Voss C, et al. Epitope-resolved serology test differentiates the clinical outcome of COVID-19 and identifies defects in antibody response in SARS-CoV-2 variants. medRxiv, 2021. doi: https://doi.org/10.1101/2021.03.16.21253716, https://www.medrxiv.org/content/10.1101/2021.03.16.21253716v1.article-info
- Peer reviewed and published scientific report.
Voss, Courtney, Sally Esmail, Xuguang Liu, Michael J. Knauer, Suzanne Ackloo, Tomonori Kaneko, Lori Lowes, et al. 2021. “Epitope-Specific Antibody Responses Differentiate COVID-19 Outcomes and Variants of Concern.” JCI Insight 6 (13). https://doi.org/10.1172/jci.insight.148855. https://insight.jci.org/articles/view/148855.