The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) across the world has caused the coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV-2, belonging to the family coronaviridae, has claimed over 2.7 million lives globally. Scientists believe that vaccination is one of the foremost approaches that can contain the ongoing pandemic. However, scientists are concerned that the new SARS-CoV-2 variants, with mutated spike proteins, may escape natural or vaccine-induced neutralizing immunity.
A study conducted in Brazil revealed the waning of antibodies against SARS-CoV-2 8 months post-infection. This report increased the need for a better understanding of the replication mechanism of SARS-CoV-2 for devising novel therapeutic strategies.
SARS-CoV-2 is a single positive-stranded RNA virus. Its genomic length is approximately 30 kb. The viral RNA is translated into the cytoplasm of infected cells by the host’s machinery. Like other viruses, SARS-CoV-2 is also dependent on cellular proteins to complete its infectious life cycle, which provides the potential means to formulate strategies to treat COVID-19.
Extensive research has been conducted to understand the molecular interactions that take place between the virus and host cell during infection. Several sophisticated techniques are used in these studies. For example, for the identification of SARS-CoV-2 virus-host protein-protein interactions (PPI), affinity purification combined with mass spectrometry is used. CRISPR-Cas9 phenotypic screens have proved to be invaluable for a better understanding of the role of host genes and cellular pathways in the life cycle of SARS-CoV-2. These studies have led to the identification of multiple potential cellular targets for drug repurposing, such as PI3K complex and the sigma-1 and -2 receptors.
Now, a new study has been published in bioRxiv* that focuses on the identification of RNA-binding protein by mass spectrometry (ChIRP-M/S) approach. This study develops a global map that contains the complete set of host proteins that involve the SARS-CoV-2 genome during infection. This study has successfully identified new SARS-CoV-2 host dependency factors by merging proteomics data with gene silencing experiments. Such a discovery has paved new roads to develop more antiviral therapies.
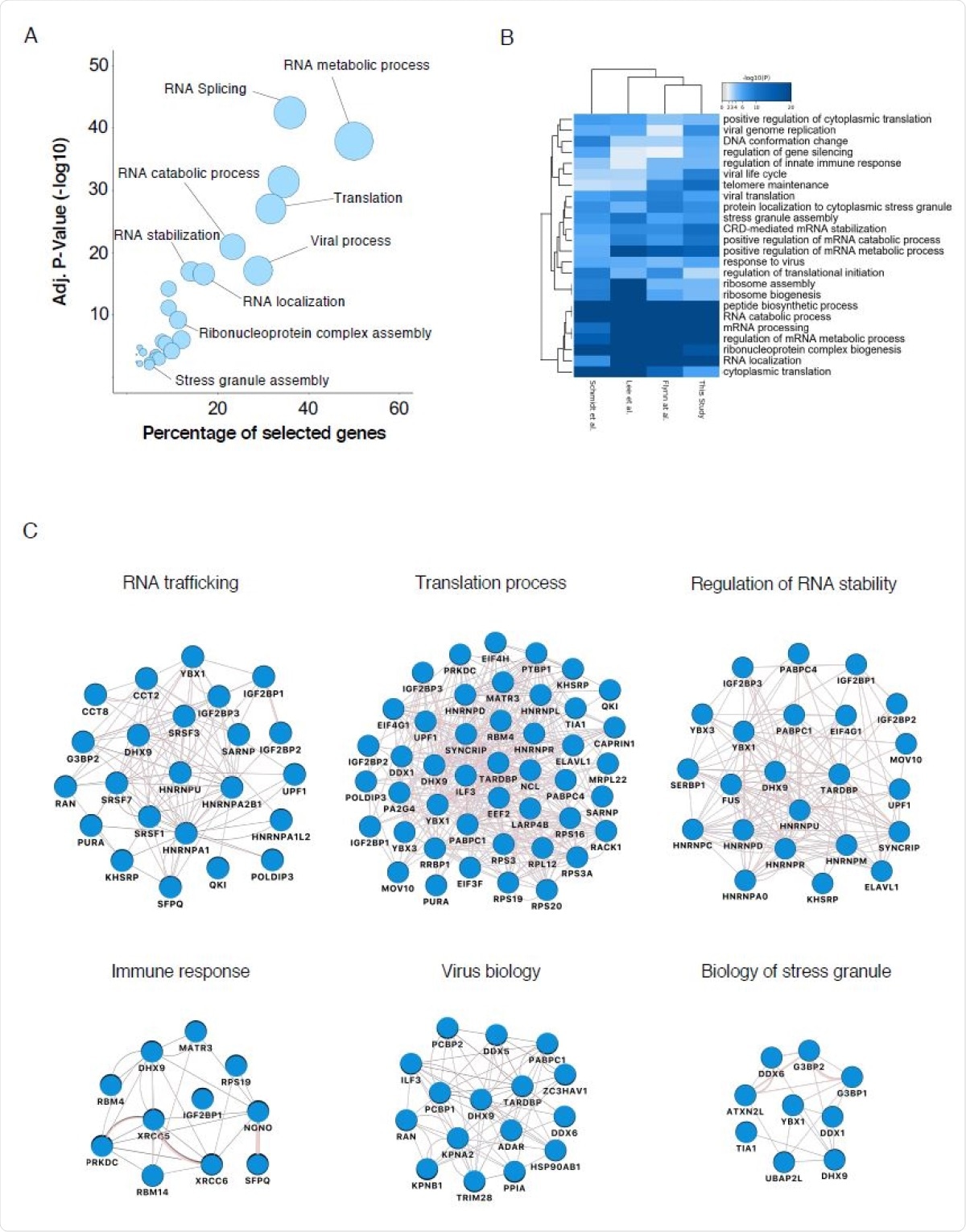
Biological analysis of the SARS-CoV-2 RNA interactome (A) GO enrichment analysis of SARS-CoV-2 RNA interactome proteins. Circles represent enriched function for an annotated ontology term and circle size correspond to the number of enriched proteins within that term. (B) Heatmap of the top-ranked GO biological processes enriched across all SARS-CoV-2 RNA interactomes (C) Selected biological process networks of SARS-CoV-2 interactome proteins.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
While characterizing the host’s factors associated with the SARS-CoV-2 genome, during infection, scientists found that the SARS-CoV-2 N protein is the most potent RNA-binding protein (RBP) of the virus. Other proteins such as structural M and S proteins, replicase polyprotein pp1ab, and the ORF9b (accessory protein) are also associated with the virus replication. ChIRP-M/S analysis revealed that among the 16 non-structural proteins (NSPs) released from pp1ab, 9 take part in the replication of coronavirus. NSP2-NSP16 has significant functions in RNA replication. Previous studies have reported that NSP1 interacts with the 40S ribosomal subunit to impede cellular translation. This protein binds with the viral 5’ untranslated region (UTR) to stimulate vRNA translation. The current research shows that the ChIRP-M/S technique can be effectively used to identify viral endogenous ribonucleoprotein (RNP) complexes that are associated with various stages (replication/transcription to particle assembly) in the SARS-CoV-2 life cycle.
The current research used SAINTexpress scoring algorithms and identified high-confident human proteins that are associated with the SARS-CoV-2 RNA. In comparison with previous studies, researchers found that around 24% to 38% of the SARS-CoV-2 RNA host factors were similar to the present data in the ChIRP-M/S interactome. Additionally, multiple RBPs associated with nucleic acid metabolisms such as splicing, mRNA stability, and transcriptional regulation poly(A) binding protein (PABP) were identified. Chaperone proteins were found to be significantly enriched and played a key role in viral protein folding. Various other components and characteristic RBPs such as paraspeckles (PS) and stress granules (SG) were among the highest-scoring candidates.
For a better understanding of the interactions between host factors and viral proteins, researchers of the current study compared the SARS-CoV-2 interactome with a reference coronavirus interactome. This study is analogous to a meta-analysis of the viral-host PPI of coronaviruses collected from 112 publications. A high-confidence hit was identified by the ChIRP assay, and the associated host proteins were known interactors of multiple coronaviruses viral proteins. The result of the current study agreed with previous studies that stated that fatty acid synthesis is essential for SARS-CoV-2 viral replication.
To understand the role of the identified RBPs in SARS-CoV-2 infection, researchers silenced their expression by RNA interference (RNAi), and thereby, identified the associated virus infection. Cathepsin L protease (CTSL) and the vacuolar ATPase subunit ATP6V1B2 were two host molecules important for SARS-CoV-2 viral entry. Further, researchers have also found that SARS-CoV2 has advanced several existing mechanisms which inhibit the type I interferon response by targeting specific RBPs.
The current research has also identified drugs that target host RBP by digging into several drug databases such as DrugBank, ChEMBL, and PanDrugs. Thereby, they have successfully identified drugs such as camptothecin (XRCC5), cerulenin (FASN), dabigatran (HNRNPC), etc., as potential antiviral agents for SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Athena Labeau, Alain Lefevre-Utile, Lucie Bonnet-Madin, Luc Fery-Simonian, Vassili Soumelis, Vincent Lotteau, Pierre-Olivier Vidalain, Ali Amara, Laurent Meertens. Characterization and functional interrogation of SARS-CoV-2 RNA interactome. bioRxiv 2021.03.23.436611; doi: https://doi.org/10.1101/2021.03.23.436611, https://www.biorxiv.org/content/10.1101/2021.03.23.436611v1
- Peer reviewed and published scientific report.
Labeau, Athéna, Luc Fery-Simonian, Alain Lefevre-Utile, Marie Pourcelot, Lucie Bonnet-Madin, Vassili Soumelis, Vincent Lotteau, Pierre-Olivier Vidalain, Ali Amara, and Laurent Meertens. 2022. “Characterization and Functional Interrogation of the SARS-CoV-2 RNA Interactome.” Cell Reports 39 (4). https://doi.org/10.1016/j.celrep.2022.110744. https://www.cell.com/cell-reports/fulltext/S2211-1247(22)00508-3.