The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to spread across the globe.
Researchers at the Institute of Medical Virology, the University of Zurich in Switzerland, have identified features in the SARS-CoV-2 genome that are important for human respiratory cell growth.
The study, published in the journal PLOS BIOLOGY, uncovered sequence features in SARS-CoV-2 variants that determine cell-specific replication. It underlines the need to monitor SARS-CoV-2 stocks carefully when phenotyping new variants of concern.
Study background
The coronavirus pandemic, which first emerged in December 2019 in Wuhan City, China, has caused over 125.43 million cases and 2.75 million deaths worldwide.
Though large-scale sequencing efforts have identified several SARS-CoV-2 variants, it is still unclear whether many of these changes impact viral replication, transmission, and adaptation.
In the virus, the Spike (S) glycoprotein stays on the surface and facilitates viral entry into the host cell by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. The viral-host membrane fusion relies on host cell proteases' cleavage, occurring after the virion's attachment to the host cell membrane or during virion maturation.
The other viral components include the Membrane (M) protein, the Nucleoprotein (N), and the Envelope (E) protein. After the virus enters into host cells, translation of viral genomic RNA causes numerous nonstructural proteins' expression. These form the replicase-transcriptase complex or fulfill additional essential functions in the virus life cycle.
Coronaviruses have a proofreading function, causing more robust genome stability compared with other RNA viruses. Many mutations in the SARS-CoV-2 genome have been reported in human circulating viruses globally. Some of these demonstrate increasing prevalence, suggesting a positive selection advantage in the new human host.
The D614G substitution in the S protein rapidly dominated sequences after it first emerged. This mutation has been tied to increased viral infectivity and replication, facilitating viral spread. Meanwhile, recently acquired mutations in the SARS-CoV-2 genome appear to promote reproduction and transmission in humans.
The study
The researchers described 14 SARS-CoV-2 isolates from patients collected during the first pandemic wave in Switzerland from March to May 2020. These isolates represent several virus clades roaming in Europe in early 2020.
However, rare additional mutations found in the S and E proteins were seen on low-passage virus stock preparation in Vero-CCL81 cells. When the researchers analyzed each SARS-CoV-2 isolate in Vero-CCL81 cells and distinguished primary human bronchial epithelial cells (BEpCs), they found the importance of the S furin cleavage site and E positions 5/6. These determine SARS-CoV-2 infectivity in human respiratory cells.
The researchers also noted that isolates that expressed S G614 appeared to replicate more effectively than isolates with the S D614.
Also, the isolates with Orf3a (Q57H) and nsp2 (T85l) substitutions showed a cell-specific replication phenotype. Isolates unique in containing these variants replicated less efficiently in Vero-CCL81 cells while maintaining efficient replication capacity in primary human BEpCs.
These variants are found in about 20 percent of SARS-CoV-2 sequences globally. Yet, their functional consequences remain unclear.
The team also observed that SARS-CoV-2 stock preparation in Vero-CCL81 cells made the isolates get passage-derived amino acid substitutions in the E protein that were not seen in patient material. They noted that SARS-CoV-2 E has crucial species- or cell type-specific functions, revealing that residues at 5 and 6 play critical roles in some vital E activities.
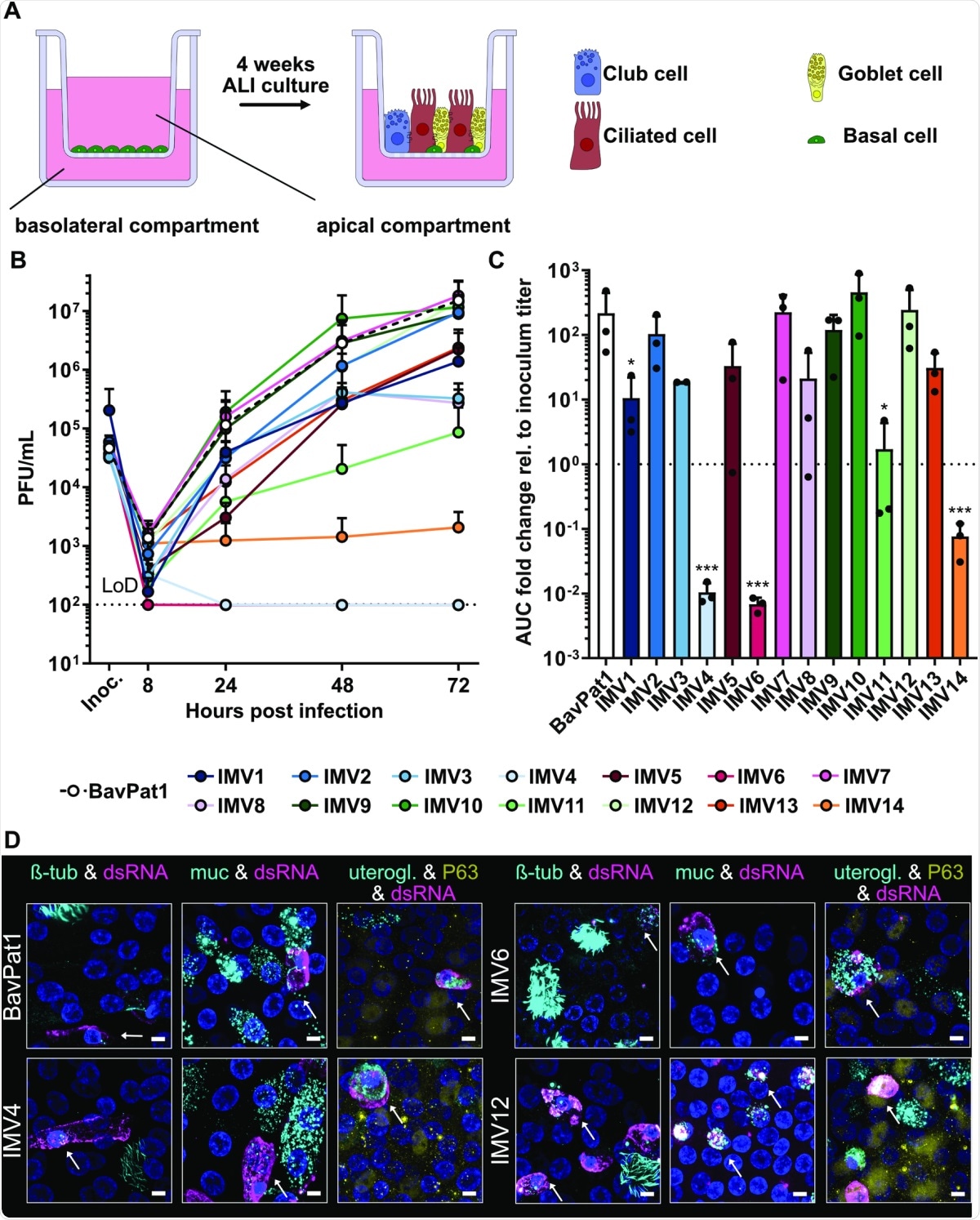
Characterization of SARS-CoV-2 isolates in primary human BEpCs.(A) Schematic representation of primary human BEpC differentiation into a pseudostratified airway epithelium by culturing at ALI on transwell plates. (B) Differentiated BEpCs from donor 1 were infected with 6,000 PFU of each SARS-CoV-2 isolate from the apical side. At several times postinfection, apical washes were harvested, and virus titers were determined by plaque assay. Data represent means and standard deviations from 3 in dependent experiments, with each experiment performed using 1 individual transwell. The dotted line crossing the y-axis at 102 PFU/mL indicates the assay LoD. (C) AUC values for the virus replication data shown in (B), normalized to inoculum titer of the respective isolate. Data represent means and standard deviations from the 3 independent experiments. A 1-way ANOVA was performed on log2-transformed data to test for significant differences between the individual isolates (p < 0.0001). To test for statistical significance against BavPat1, an unpaired t test was used on log2-transformed data (*p < 0.05; ***p < 0.001). (D) Indirect immunofluorescent staining of SARS-CoV-2–infected BEpCs. Differentiated BEpCs were infected from the apical side with 6,000 PFU of the indicated SARS-CoV-2 isolate. At 72 h postinfection, cells were fixed, permeabilized, and stained for the presence of infected cells (dsRNA; magenta) and ciliated cells (β-tubulin; cyan), goblet cells (Muc5AC; cyan), club cells (uteroglobin; cyan), or basal cells (P63; yellow). Nuclei were stained with DAPI (blue). Z-stacks were transformed into maximum projection images. Scale bar represents 9.6 μm. Arrows indicate co-localization. Data show a subset of representative images from 2 independent experiments performed in different BEpC donors (see also S5–S10 Figs). For underlying data, see S1 Data. ALI, air–liquid interface; AUC, area under the curve; BEpC, bronchial epithelial cell; LoD, limit of detection; PFU, plaque-forming unit; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2.
The team also found that in-frame deletions of the furin cleavage site in S could be enriched in SARS-CoV-2 isolates during passage in Vero-CCL81 cells. Vero cells appear to lack TMPRSS2 expression. This means that maintaining the S furin cleavage site is unlikely to benefit SARS-CoV-2 in these cells.
These findings emphasize the need for care during stock preparation, specifically where viruses are being compared with each other for phenotypes. The team added that different cell lines like the human Calu-3 are more appropriate for SARS-CoV-2 propagation.
Yet, the team found that in-frame deletions in the S furin cleavage site weakened the SARS-CoV-2 virus in BEpCs.
"Our study of SARS-CoV-2 cell adaptations revealed the critical nature of this S furin cleavage site for SARS-CoV-2 replication in primary human respiratory cells, a finding similar to that recently described by others using alternative methods," the researchers noted in the study.
They concluded that the findings describe vital features in the SARS-CoV-2 genome that are important for viral growth in human respiratory cells.
Understanding how SARS-CoV-2 variants affect cells is crucial for scientists to develop effective therapies and vaccines to curb the pandemic.