A study conducted at the Los Alamos National Laboratory, USA, and Concordia University, Canada, has explored the impact of different types of glycans on the interaction between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein and human angiotensin-converting enzyme 2 (ACE2) receptor.
The findings reveal that although the spike – ACE2 interaction is not directly influenced by the glycans, there are specific glycans on ACE2, such as mannose-9 (MAN9) and sialated complex flucosylated 2-antennae (FA2), which can significantly alter the affinity between spike protein and ACE2. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly infectious RNA virus responsible for the current coronavirus disease 2019 (COVID-19) pandemic. It is now well-established that SARS-CoV-2 infection initiates with the interaction between spike receptor-binding domain (RBD) and host cell ACE2 receptor. This is followed by proteolytic activation of the viral spike and subsequent fusion between the virus envelop and host cell membrane.
Structurally, both spike protein and ACE2 are densely glycosylated with distinct types of asparagine-linked N-glycans. Some of these glycans are present in the RBD-ACE2 interface, suggesting that glycans may influence the RBD-ACE2 binding characteristics. Previously, it has been observed that glycans play a role in modulating epitope exposure and antibody affinity for spike RBD. Moreover, there is evidence indicating that the host cell entry of SARS-CoV can be prevented by disrupting ACE2 glycosylation.
In the current study, the scientists have explored whether different types of glycans play any role in modulating spike RBD – ACE2 interaction. They have used molecular dynamics simulation approaches to analyze their study questions.
Study design
In the study, the scientists analyzed two models of RBD – ACE2 complex. In one RBD – ACE2 model, MAN9 glycans were attached to the Asn53, Asn90, Asn103, 59 Asn322, Asn432, and Asn546 or b residues of ACE2. Similarly, in another model, FA2 glycans were attached to the same ACE2 residues. In both models, a single FA2 glycan was attached to the Asn343 of spike RBD.
To evaluate the impact of glycans, they conducted all-atom explicit-solvent simulations of three conditions: the RBD – ACE2 complex with MAN9 glycans; the RBD – ACE2 complex with FA2 glycans; and the RBD – ACE2 complex without any glycan.
Important observations
The simulation study findings revealed that the physical contact between spike RBD and ACE2 was not affected by the presence of glycans and that the glycans did not influence relative orientations of RBD – ACE2. Moreover, the simulation data identified that interactions between the residues in the ACE2 N-terminal helix and the receptor-binding motif of RBD formed the most persistent complexes.
In another set of experiments, the scientists studied the effect of N501Y mutation on the RBD – ACE2 interaction. The N501Y is a spike RBD mutation primarily observed in the newly emerged UK variant (B.1.1.7) of SARS-CoV-2. By analyzing the simulated interactions, they observed that the N501Y RBD mutation further stabilized the RBD – ACE2 complex by interacting with Y41 and K353 of ACE2. This explains the increased transmissibility observed in the UK variant of SARS-CoV-2.
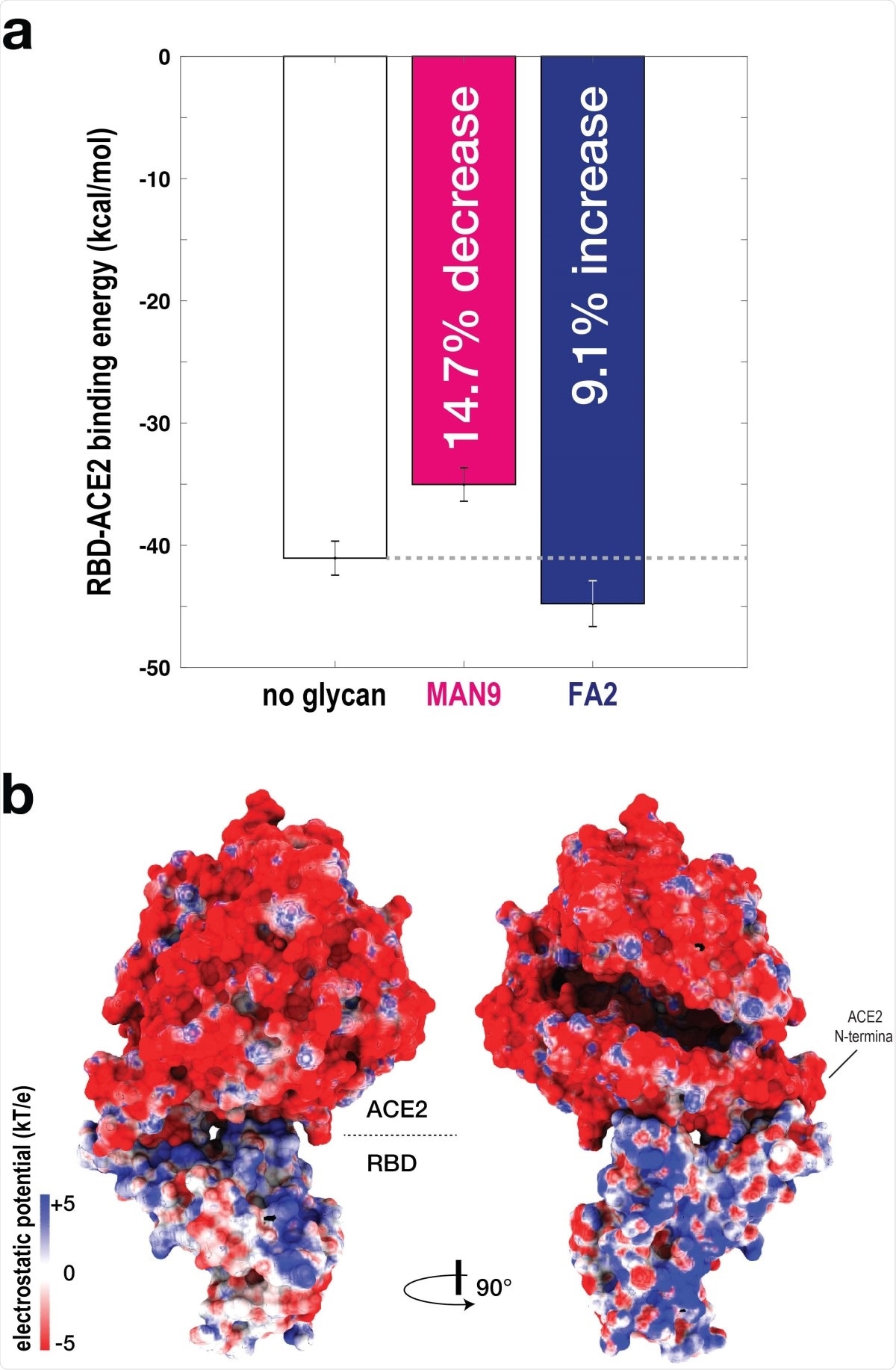
RBD-ACE2 binding affinity is glycan dependent. (a) To evaluate the binding energy between RBD-ACE2, the MM-PBSA approach was applied. Binding energy was calculated for simulations without glycans (white bar), and with MAN9 glycans (magenta bar) or FA2 glycans (blue bar) on ACE2. Simulations with MAN9 glycans on ACE2 are associated with a 14:7% decrease in stability, relative to the non-glycan simulations. In contrast, simulations with FA2 glycans on ACE2 result in a 9:1% increase in stability. (b) Electrostatic surface potential calculated for ACE2 and RBD. Two complementary views show that the ACE2 surface is overall negatively charged, while the surface of RBD is overall positive.
To further examine the RBD – ACE2 glycan interactions, the scientists thoroughly analyzed the interactions between RBD residues and MAN9 or FA2 glycan attached to ACE2. Their findings revealed that the probability of interaction between the spike RBD and the glycans present on Asn90 residue of ACE2 was significantly higher than that of other glycans. However, they noticed that the contact between RBD and Asn90 ACE2 glycan did not compete with the RBD – ACE2 interaction at the protein level. With further analysis, they observed that the steric effects of Asn90 glycan delayed the dissociation of RBD, which in turn increased the stabilization of RBD – ACE2 interaction.
Furthermore, they investigated the impact of glycan – glycan interactions on the RBD – ACE2 binding affinity. The findings revealed that Asn343 glycan of RBD more frequently interacted with Asn53 and Asn322 glycans of ACE2, suggesting that these interactions may contribute to RBD – ACE2 affinity.
To investigate relative alterations in RBD – ACE2 interactions with and without glycans, they performed a binding energetics analysis using the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) approach. The findings revealed that while MAN9 glycan on ACE2 reduced the RBD – ACE2 affinity, the presence of FA2 glycan increased the affinity. Furthermore, the findings identified that distinct electrostatic properties of the negatively charged sialic acids in FA2 glycan are the main contributors of higher affinity.
Study significance
The study highlights the importance of specific glycan types in modulating the binding affinity between spike RBD and ACE2. In other words, different types of glycans attached to the host ACE2 receptor determine the stability of RBD – ACE2 interaction. Interestingly, the findings reveal that the electrostatic properties of specific ACE2 glycans influence the binding affinity without altering the physical interactions between virus and host.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources