A team of international scientists has recently explored the transmission dynamics of currently circulating variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a southern African country. By analyzing the genomic sequences of these variants, they have identified a novel SARS-CoV-2 variant with multiple spike mutations. They have temporarily designated the variant A.VOI.V2. A detailed description of the genomic surveillance they carried out is currently available on the medRxiv* preprint server.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
Since its emergence in December 2019, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has undergone more than 12,000 mutations. The majority of these mutations are found in the viral spike protein, which is a glycoprotein on the viral envelope required for establishing SARS-CoV-2 infection. Because of robust immunogenicity, the spike protein is considered to be the most potent target for developing therapeutic antibodies and vaccines. An increased frequency of spike mutations in newly emerging variants has raised the question of whether these mutations could potentially impair the functionality of monoclonal antibodies and vaccines designed specifically against the previously circulating viral variants.
Some recently emerged SARS-CoV-2 variants of concern (VOCs), such as the UK, South African, and Brazil variants, have shown significantly higher transmissibility than the previously circulating variants. Moreover, a growing pool of preliminary studies has suggested that these variants might be associated with increased virulence. The mutations found in these variants have been found to increase the binding affinity between spike receptor-binding domain (RBD) and host angiotensin-converting enzyme 2 (ACE2) receptor. Furthermore, there is evidence indicating the some of the spike RBD mutations facilitate the virus to escape antibody-mediated neutralization. These observations explain the reason behind the increased infectivity and virulence of VOCs.
In the current study, the scientists have conducted thorough genomic surveillance of SARS-CoV-2 variants across Angola, a country located on the west coast of southern Africa. The study has been conducted by the Network for Genomic Surveillance in South Africa (NGS-SA) in association with the Africa Centers for Disease Control and Prevention and the African Society of Laboratory Medicine.
Study design
Soon after its emergence at the end of 2020, the South African variant of SARS-CoV-2 (lineage B.1.351) has spread rapidly to over 50 countries globally and become the predominantly circulating strain in southern Africa. The current study has been initiated to rapidly characterize the transmission dynamics of this variant and other VOCs in Africa. The first report on the SARS-CoV-2 genomic surveillance data from Angola has been presented here.
Important observations
The scientists analyzed a total of 118 nasopharyngeal samples collected between June 2020 and February 2021. Using these samples, they generated 73 high-quality SARS-CoV-2 genomes; of which, 14 were from B.1.351, B.1.1.7, and B.1.525 lineages; 44 were from C.16 lineage from Portugal; and 12 from other lineages. In addition, they identified a novel variant in three air passengers from Tanzania. Interestingly, three viral genomes isolated from these passengers displayed almost identical sequence. They temporarily designated the novel variant A.VOI.V2.
With further analysis, they observed that the novel variant has a total of 31 amino acid substitution mutations and 3 deletion mutations. Of these mutations, 11 out of 31 substitution mutations and all deletion mutations were found in the spike protein. By specifically analyzing the spike mutations, they identified 3 substitutions in the spike RBD, 2 substitutions nearby S1/S2 cleavage site, and 5 substitutions and 3 deletions in the spike N-terminal domain (NTD). Moreover, they observed that some of the NTD mutations are present in the antigenic supersite.
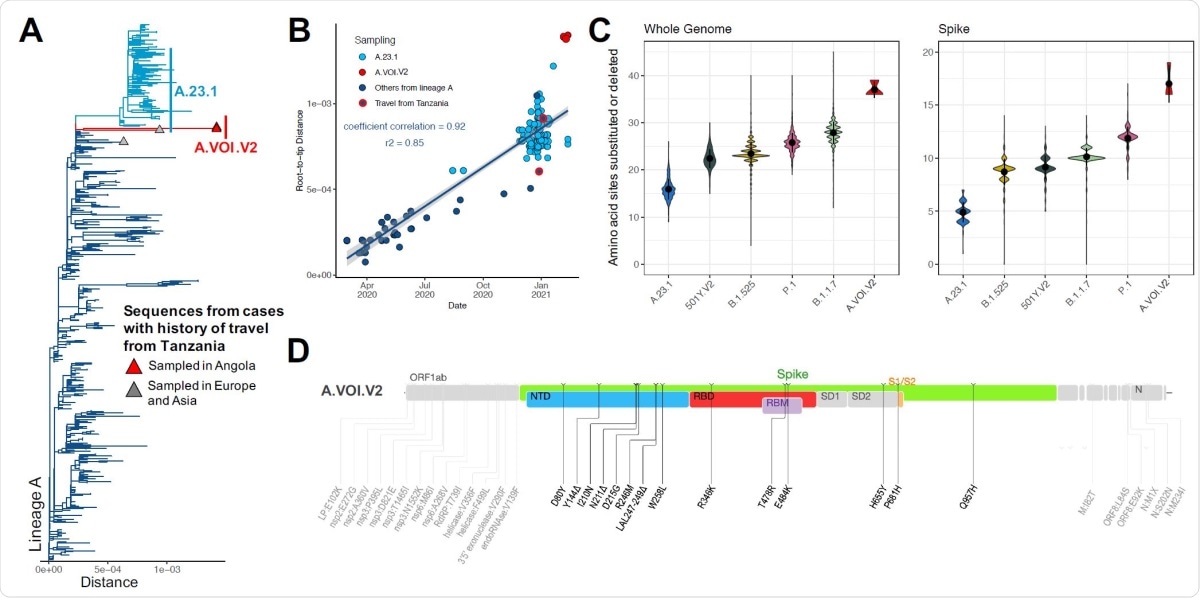
A) Phylogenetic tree of a subset of lineage A sequences (n=319) including five sequences from cases with history of travel of Tanzania, three of which are the A.VOI.V2 sampled in Angola (tips shown with a triangle); B) Regression of root-to-tip genetic distances against sampling dates, for sequences belonging to lineage A, showing the novel A.VOI.V2 (red), the known VOI A.23.1 (light blue), other sequences of lineage A (deep blue), two of which are documented to have travel history from Tanzania (red outline); C) Violin plot showing the number of amino acid mutations in the whole genome and spike glycoprotein in a subset of genomes from five known variants compared to the novel A.VOI.V2; D) Genome map showing the position of the 31 amino acid substitutions and three deletions (spike in color, NTD = Nterminal domain, RBD = receptor-binding domain, RBM = receptor-binding motif, S1/S2 = S1/S2 cleavage site, and the rest of the genome in grey).
Study significance
The study identifies a novel SARS-CoV-2 variant with multiple spike mutations. The majority of these mutations are also present in other VOCs and are known to increase viral infectivity and antibody resistance. These observations indicate that in response to certain selective pressures, these mutations are gradually evolving under positive selection and improving viral fitness.
Although the novel variant is identified only in three passengers from Tanzania, the scientists believe that more investigations are urgently needed to control its transmission within and out of the source country.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.