With the coronavirus disease 2019 (COVID-19) pandemic continuing around the globe, new mutants of SARS-CoV-2 are emerging. These new variants are likely more infectious and can better evade our immune response.
The SARS-CoV-2 spike protein, in particular the RBD, is key in binding to host receptors, mainly the angiotensin-converting enzyme 2 (ACE2) in humans. One highly conserved region of the RBD, called antigenic site II, can elicit neutralizing antibodies. However, this region is generally inaccessible because of the RBD conformation, and there is a low fraction of antibodies targeting this site in infected individuals.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In a new study published in the bioRxiv* preprint server, researchers report a new monoclonal antibody that is targeted toward site II and has a broad neutralizing capability.
Testing potency of monoclonal antibody
The authors sorted spike protein-specific memory B cells from a convalescent individual 75 days after symptom onset. They found one monoclonal antibody, called S2X259, which reacted with 29 of 30 spike proteins of sarbecoviruses, including SARS-CoV-2 and its new variants. The antibody also reacted with bat sarbecoviruses, suggesting its broad neutralizing capability.
The antibody also bound strongly to 10 RBDs from different sarbecoviruses. The binding of this antibody was not affected by the different single-point RBD mutations seen in the new variants of SARS-CoV-2, including the United Kingdom, South African, Brazilian, and the B.1.427/B.1.429 variants.
Using pseudotyped virus systems, the team found that the antibody neutralized SARS-CoV-2 and did not lose its potency against the different variants or the N439K or Y453F mutation. The antibody not only neutralized a variety of sarbecoviruses that use the ACE2 receptor but also cross-reacts with sarbecoviruses that do not use ACE2 for infection.
To understand how this antibody has high neutralizing potency, the team imaged the complex formed between the spike protein and the antibody using cryo-electron microscopy. They found that the antibody recognizes a glycan-free site, which requires two RBDs to be in the open conformation. It forms contacts with residues 369-386, 404-411, and 499-508 in the RBD.
The epitope the antibody binds to is conserved in all the circulating SARS-CoV-2 variants. In addition, it does not target the 417 or 484 residues (mutations here are found in B.1.351 and P.1), and this could be why it is potent against the different variants.
The action of this antibody does not affect the neutralization effect of class 1 and class 3 antibodies. The majority of approved antibodies for clinical use belong to these classes. Hence, the new antibody can be used in combination with other antibodies to increase neutralization breadth.
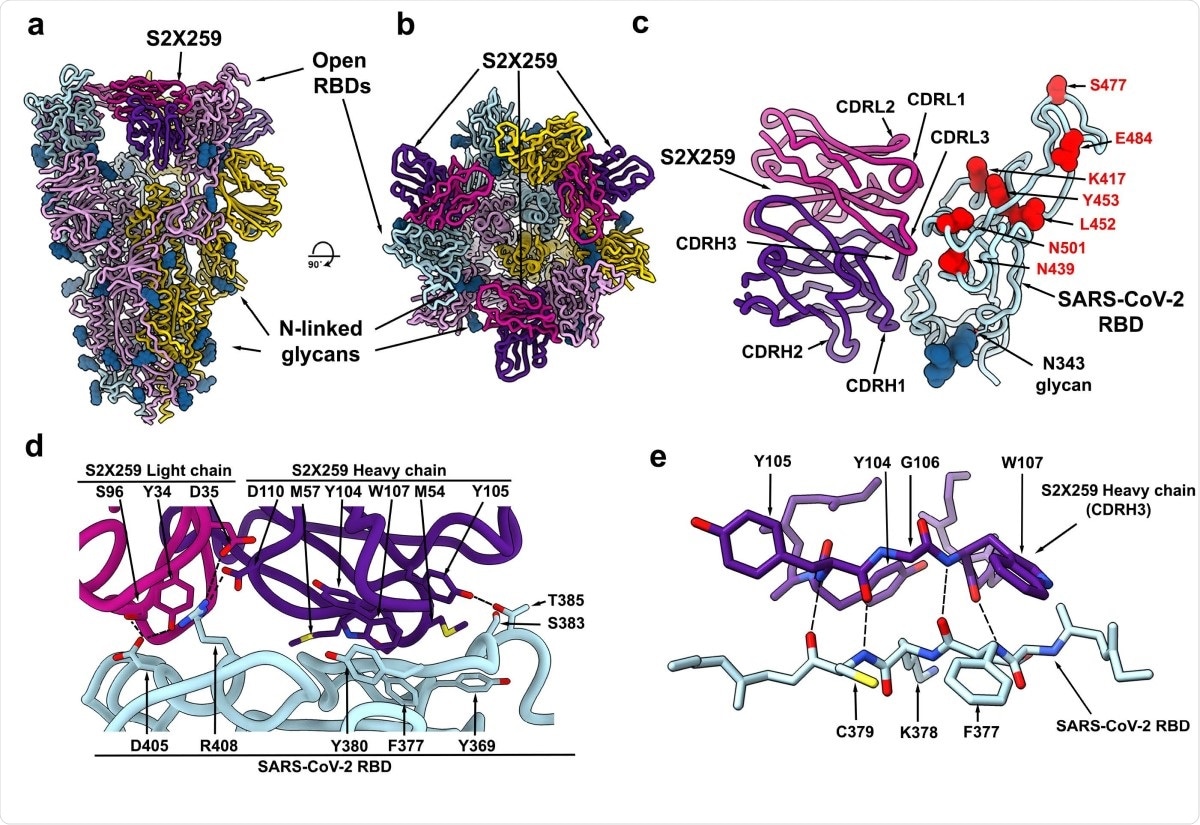
The S2X259 broadly neutralizing sarbecovirus mAb recognizes RBD antigenic site II. a-b, CryoEM structure of the prefusion SARS-CoV-2 S ectodomain trimer with three S2X259 Fab fragments bound to three open RBDs viewed along two orthogonal orientations. c. The S2X259 binding pose involving contacts with multiple RBD regions. Residues corresponding to prevalent RBD mutations are shown as red spheres. d-e, Close-up views showing selected interactions formed between S2X259 and the SARS-CoV-2 RBD. In panels a-e, each SARS-CoV-2 S protomer is coloured distinctly (cyan, pink and gold) whereas the S2X259 light and heavy chain variable domains are coloured magenta and purple, respectively. N-linked glycans are rendered as blue spheres in panels a-c.
Potential use against a broad range of sarbecoviruses
Using computational analysis, the team determined what RBD mutations could escape the antibody from binding. They found only a few RBD mutations disrupt the binding of this antibody. The substitution of the residue at position 504 gave the most significant disruption in binding.
When they replicated a pseudotyped SARS-CoV-2 virus in the presence of the S2X259 antibody, the only mutation they found caused by selective pressure was G504D. This mutation has rarely been seen in human isolates so far.
The selection of a single escape mutation suggests the region targeted by the antibody might not tolerate amino acid substitutions without changing viral fitness. Hence it is conserved across different sarbecoviruses. Thus, there is a high barrier for the emergence of mutations against this antibody, suggesting it could become key in combating the pandemic.
When Syrian hamsters were challenged with SARS-CoV-2, with the antibody administered 48 hours before virus infection, the authors found more than two orders of magnitude decrease in virus in the lungs compared to hamsters that did not receive any treatment. In addition, the antibody also protected hamsters infected with the B.1.351 strain.
The detection of a large variety of sarbecoviruses in bats and other mammals, along with the increased human-animal interactions, makes it likely that more cross-species transmission of viruses can occur. With increasing evidence that antibodies targeting the RBD form a major proportion of neutralizing activity, RBD-based vaccines could elicit high levels of antibodies like S2X259 with high potency. Such strategies can help overcome the current COVID-19 pandemic and help prepare for future sarbecovirus infections.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Tortorici, M. A. et al. (2021) Structural basis for broad sarbecovirus neutralization by a human monoclonal antibody. bioRxiv. https://doi.org/10.1101/2021.04.07.438818, https://www.biorxiv.org/content/10.1101/2021.04.07.438818v1
- Peer reviewed and published scientific report.
Tortorici, M. Alejandra, Nadine Czudnochowski, Tyler N. Starr, Roberta Marzi, Alexandra C. Walls, Fabrizia Zatta, John E. Bowen, et al. 2021. “Broad Sarbecovirus Neutralization by a Human Monoclonal Antibody.” Nature 597 (7874): 103–8. https://doi.org/10.1038/s41586-021-03817-4. https://www.nature.com/articles/s41586-021-03817-4.