The current pandemic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has triggered an intensive hunt for effective new and repurposed antivirals and antibodies, for prevention and treatment of the infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Spike-targeting neutralizing antibodies
SARS-CoV-2 possesses multiple spikes on its surface, which mediate its attachment to the target host cell receptor, the angiotensin-converting enzyme 2 (ACE2), and viral entry into the cell. The spike-receptor interactions mostly take place at the receptor-binding domain (RBD) on the S1 subunit of the spike.
The extent of protection from infection in the case of SARS-CoV-2 is probably linked to the neutralizing antibody titer. Those with the most potent neutralizing ability are directed against the RBD and are found to predominate in polyclonal sera from convalescent COVID-19 patients and in COVID-19 vaccine recipients, as well as in therapeutic antibodies.
The recently emerged South African (SA) and Brazil variants show varying resistance levels to vaccine-induced neutralizing antibodies. These are characterized by mutations affecting the RBD, which impact its binding to such antibodies.
Study aims
The current study seeks to explore both the potential existence of such antibodies, as well as to find antibodies that have broad activity against SARS-related coronaviruses (Sarbecoviruses). These would be essential, given the future potential for outbreaks of other Sarbecoviruses.
The researchers in this study worked on a previously reported antibody S309, which triggers potent effector activity and has neutralizing activity against currently circulating SARS-CoV-2 isolates as well as SARS-CoV. This was developed as the therapeutic antibody VIR-7831, which shows clinical efficacy in treating COVID-19.
The attempt is to better understand how the RBD antibody-binding sites or epitopes are related to the potency of neutralizing antibodies, resistance to viral evasion mutations, and the cross-reactivity of such antibodies to other Sarbecoviruses.
The researchers made use of a panel of different antibodies using binding assays, deep mutational scanning, positive selection of viral escape, and the biochemical and structural characteristics of such antibodies. Six of these antibodies were reported for the first time in this study.
The antibodies studied here bound a variety of epitopes, including those that overlap the receptor-binding motif (RBM) in the RBD, and those within the RBM itself, and show a range of RBD-binding affinities and neutralizing potencies against both wildtype and pseudoviruses particles expressing the SARS-CoV-2 spike antigen.
Effect of size of escape mutation
There are many different sites of mutational escape, depending on the antibody. With narrow functional epitopes (a sequence that is crucial to antibody binding), a single or a few key mutations could abolish RBD binding. One such antibody is S309. Others, such as S2H13, have broad functional epitopes.
Mapping escape mutations
In order to predict the effects of such mutations, the investigators had previously assessed the effect of all RBD mutations on the binding affinity to the ACE2 receptor, and on the higher structure of the protein – which determines its function.
These effects were combined to produce a computed measure of how easy it is to escape neutralization by each antibody – an escape mutation map. This, in turn, reflects viral tolerance for such functional mutations.
The study also looked into the sensitivity of each antibody to mutations reported in the GISAID (Global Initiative for Sharing All Influenza Data) database up to March 4, 2021.
The researchers found that the impact of natural mutations in the viral RBD was much greater for some antibodies than for others. These include mutations found in fast-spreading viral strains, called the variants of concern (VOCs).
They also found that of the four antibodies that bind to the core RBD, cross-reactivity and tight binding were observed with SARS-CoV and other ACE2-binding Sarbecoviruses with over 70% amino acid identity in the RBD.
In addition, S304 and S2H97 showed the ability to cross-bind other more different non-ACE2-binding RBDs with only 64% identity to the current virus.
“S2H97 [is] the broadest pan-sarbecovirus RBD antibody described to date.” Pseudovirus assays confirmed that S2H97 neutralizes a variety of different spike variants from both SARS-CoV-2- and SARS-CoV.
Antibodies that bind only RBM-restricted epitopes show much lower cross-binding, involving only the RBD of the SARS-CoV-2 and the pangolin CoV.
A unique finding was that S2E12 binds to the RBD of all SARS-CoV-2 variants and to the pangolin CoV, which has only 86% identity with the former. This suggests that some functional epitopes tolerate more substitutions than others within the constantly evolving RBM.
Structural basis of neutralization potency
The earlier escape mutation mapping was used to understand how similarly binding escape mutations look in two dimensions. It shows how epitopes move across the RBM in a smooth progression, beginning from that binding to the Lilly antibody LY-CoV016 through S2X58 to that produced by Regeneron, REGN10987.
The 2-D projection also shows which antibodies bind to unique parts of the RBD. For instance, S2H97 engages with the side of the core RBD, below the receptor-binding ridge, adjacent to the N-terminal domain (NTD) of the neighboring protomer of the trimeric spike protein, in the closed configuration.
Connecting the in vitro neutralization potency of various antibodies, their binding spectrum and immune evasion properties with this projection allowed an exploration of their structural basis.
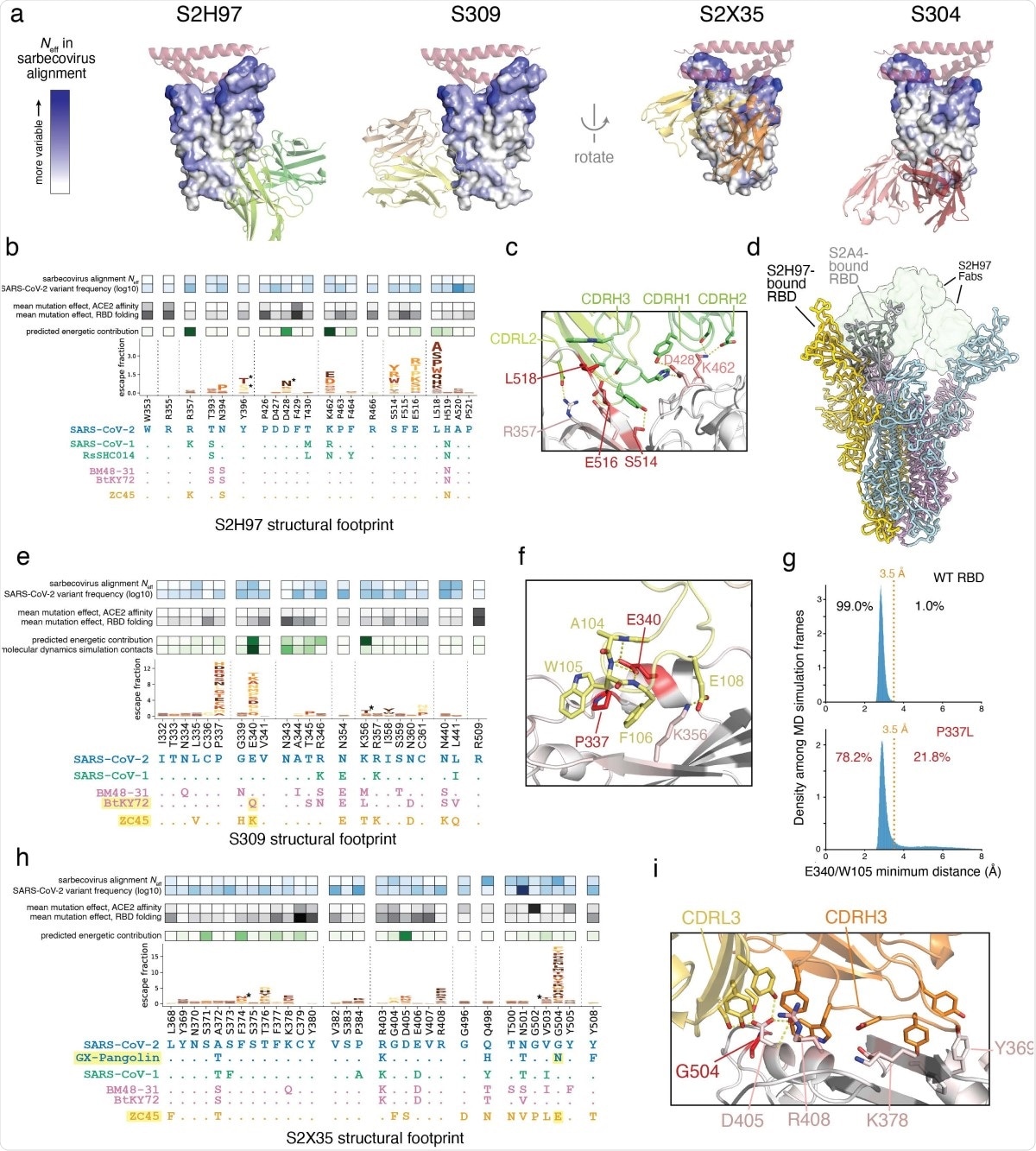
Structural basis for broad sarbecovirus binding. a, Overview of the surfaces targeted by broadly binding RBD antibodies. RBD surface is colored by site variability in the sarbecovirus alignment (effective number of amino acids, scale bar at left). ACE2 (key motifs) shown in transparent cartoon. Antibody variable domains shown as cartoon, with darker shade indicating the heavy chain. b, Integrative genetic and structural features of the S2H97 structural epitope (5 Å cutoff). Heatmap details and scale bars as in Fig. 3e,f. Logoplots are colored by mutation effects on folded RBD expression (see scale bar, Fig. 1b). Asterisks in logoplot indicate escape mutations that introduce N-linked glycosylation motifs (NxS/T). Below the logoplot is a selection of aligned sarbecovirus RBDs (sequenced colored by clade as in Fig. 1d, Extended Data Fig. 5a). c, Zoomed in view of the S2H97/RBD interface, with important contact and escape residues labeled. d, CryoEM structure of S2H97-bound SARS-CoV-2 S. Spike protomers are shown in yellow, blue, and pink, and S2H97 Fabs in transparent green surface. S2A4-bound spike protomer from PDB 7JVC17 is shown in gray and aligned to the yellow subunit, indicating the additional extent of RBD opening necessary to access the S2H97 epitope compared to a class II antibody. e, Integrative features of the S309 structural epitope, details as in (b). An additional row in the heatmap overlay reflects the proportion of all close S309/RBD contacts (<3.5 Å) made by each residue during molecular dynamics simulation. Highlighted sarbecoviruses identify those that escape S309 binding, and highlighted mutation in the alignment is the likely contributor according to our escape map. f, Zoomed in view of the S309/RBD interface, with important contact and escape residues labeled. g, Molecular dynamics simulations of the S309/RBD structure. Histograms show the distribution of minimum distance between E340RBD and W105HC heavy atoms across 1-ns frames during the simulation of the unmutated (top, 42-µs simulation) and P337L mutated (bottom, 91-µs simulation) RBD bound to S309. Orange line reflects the 3.5 Å distance cutoff used to define close contact. Percentage of frames in which E340 and W105 are or are not in close contact is labeled. See Extended Data Fig. 8c for the occupancy of other S309:RBD contacts across the simulations. h, Integrative features of the S2X35 structural epitope, details as in (b). i, Zoomed in view of the S2X35/RBD interface, with important contact and escape residues labeled.
Antibodies with the greatest neutralizing ability had the narrowest functional epitopes, binding within the RBM epitopes.
Conversely, those binding to the core RBD epitopes showed lower levels of neutralization. Of course, neutralization is not the only mechanism of protection against infection for RBD-binding antibodies.
Antibodies with broad sarbecovirus binding universally target the core RBD, illustrating a general tradeoff between breadth of sarbecovirus binding and potency of SARS-CoV-2 neutralization.”
Location and immune evasion
The number of RBD escape mutants within the functional epitope of each antibody (its “size”) is not closely related to its location. Instead, narrower functional epitopes, with fewer escape mutations, have higher binding affinity between the Fab (antigen-binding fragment) and the RBD.
However, the location does affect the link between the size of the functional epitope and its potential for immune escape. The S2E12 and the S2X58 antibodies have similar functional epitope breadth, unlike the narrower functional epitope of S2D106.
S2E12, however, binds to crucial ACE2-contacting amino acids, where mutations would impair the secondary folding of the RBD, abolishing its function. Few such escape mutations have been observed.
S2E12 can bind multiple Sarbecoviruses showing significant identity to SARS-CoV-2, including the bat RaTG13 and pangolin CoVs. Indeed, a broader range of binding among core RBD antibodies indicates high affinity binding and low escape breadth, since they bind to sites with functional constraints.
Conversely, S2X58 and S2D106 bind to epitopes that can vary across different SARS-CoV-2 variants, indicating functional tolerance.
As expected, of the seven antibodies tested for positive selection of the escape mutants, the use of S2X58 led to the emergence of viral variants with multiple scattered mutations. S2D106 allowed only mutations at two sites.
Escape mutations selected by these antibodies include L452R and E484K, as found in the VOCs.
However, S2E12 selects for mutations at few sites associated with low ACE2 binding affinity. These mutations, being functionally constrained as well as under less selection pressure by natural antibodies in polyclonal sera, are not found in VOCs.
Structural basis for escape mutations
The reason for this difference is structural. S2E12 binding to the RBD core causes packing of RBD F486 and N487 residues within a buried hydrophobic cavity, at the junction of antibody heavy and light chains. The first residue makes contact with the ACE2 receptor, while the second forms polar contacts with the antibody backbone.
In contrast, S2E12 binding sites such as E484 and S477, at the periphery of the ACE2-RBD interface, have more variation in sequence, being less functionally constrained. This explains why S2E12 binds with high affinity to common mutants at these sites.
It also explains its binding to RaTG13 and pangolin CoV, with the F486L mutation that allows similar packing in the same hydrophobic cavity.
With S2D106 binding, the antibody is tethered by a buried polar contact between RBD E484 and antibody R96 residues, and by nonpolar contacts between the RBD F490 and the heavy chain complementarity domain region 2 (CDR2) of the antibody.
With no indispensable role in binding, the E484 and F490 residues can easily harbor escape mutations while retaining their function.
This comparison highlights how a subtle change in the focus of the RBD: antibody interaction impacts the robustness of each antibody to viral escape, including among natural SARS-CoV-2 mutants.”
Structural impact for antibody breadth
The study also shows that S2H97 is so broad in its binding across Sarbecoviruses because it binds to a hitherto unrecognized highly conserved site V. During this binding, the heavy chain CDR3 is packed into a crevice of the RBD occurring at the center of the epitope.
Additionally, all three heavy chain CDR3 loops and the light chain CDR2 make crucial polar interactions with the RBD, making this a functionally constrained binding surface.
Mutations here impair RBD folding, especially with quaternary NTD packing in the closed state of the trimeric spike. Moreover, this binding surface is open only with a widely opened RBD. This explains the scarcity of antibodies to this site and the low neutralization potency of S2H97, despite its high affinity.
S309 binding binds to a small area of the RBD and thus does not permit significant diversity of structure. The escape mutations here involve E340 and P337 residues, with electrostatic and van der Waals interactions with CDRH3.
The former residue is key to antibody contact. Mutations at the P337 residue allow escape by steric hindrance or by loss of stability of E340.
The narrow binding epitope means that S309 neutralizes SARS-CoV-2 mutants in current circulation and also binds RBDs of other Sarbecoviruses that have a conserved E340, despite having eight different residues within the binding area of this antibody.
S2X35 has a large binding interface, but escape here is not because of changes in the side chains that are necessary for antibody binding. Instead, the G504 residue, when mutated, creates steric hindrances with D405 and the antibody’s light chain. D405 mutation entails very drastic changes in the sequence. This low potential for escape indicates no single RBD side chain is responsible for its binding to the antibody, which in turn allows broad sarbecovirus binding.
What are the implications?
Rather than a single-minded quest for increased potency of neutralization, important as this aspect is, these results indicate that “screening antibodies for high-affinity binding and sarbecovirus breadth identifies antibodies that are robust to ongoing SARS-CoV-2 evolution.”
As VOCs emerge, vaccine-elicited antibody resistance is a major concern. Most VOC mutations are in the RBM, including E484, K417 and L452. These prevent binding by polyclonal serum and certain therapeutic monoclonal antibodies. Five antibodies in this panel, targeting the RBM, could be evaded by mutations at position 484.
The use of escape maps could help identify antibodies that resist such immune escape and the selection of therapeutic antibodies against the variants in circulation at the time in that locality.
The future may include newly dominant mutations, including some in the RBM that prevents antibody binding. Thus, antibodies that recognize broad epitopes should be focused on, as well as epitopes that are highly conserved, including residue-level differences among the various epitopes. This will help identify and implement better interventions that resist such immune escape both in the current pandemic and in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources