A team of scientists from the United Kingdom recently investigated the ultrastructural details of the attachment, entry, and budding processes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the human airway epithelium. They have used a highly differentiated air-liquid interface cultures of airway epithelium to thoroughly investigate the viral infection cycle. The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), is an enveloped RNA virus belonging to the Coronaviridae family. The virus primarily attacks human airway epithelial cells to initiate infection. Mechanistically, the receptor-binding domain (RBD) of the S1 subunit of the viral spike glycoprotein binds to angiotensin-converting enzyme 2 (ACE2), which is ubiquitously expressed at the apical surface of host airway epithelial cells. This is followed by proteolytic cleavage of the spike S1/S2 subunit by a serine protease TMPRSS2 present within airway epithelial cells. This leads to dissociation of the S1 subunit and a conformational alteration in the S2 subunit to generate a post-fusion state of the spike protein, which in turn is necessary for the fusion of the viral envelope to the epithelial cell plasma membrane. Upon fusion, the viral components are released from the viral lumen into the host cell cytoplasm to initiate viral replication.
Evidence suggests that viral replication occurs at the perinuclear sites where endoplasmic reticulum-derived double-membrane vesicles are formed. The double-stranded RNA-enriched vesicles are associated with viral replication. Although there is evidence to suggest that the budding of SARS-CoV-2 occurs at endoplasmic reticulum-to-Golgi intermediate compartments, the exact route from double-membrane vesicle to viral budding to release of the virion from cells is still not fully known.
In the current study, the scientists have investigated the ultrastructure of human airway epithelial cells infected with three SARS-CoV-2 variants, including B.1.1.7, B.1.258, and B.1.117.19.
Study design
Using various cell culture methods, the scientists developed highly differentiated air-liquid interface cultures of human airway epithelial cells collected from nasal brushing samples of healthy and SARS-CoV-2-infected individuals. In other words, by differentiating epithelial cells at an air-liquid interface, they developed an epithelial layer containing ciliated, goblet, and basal cells.
After infecting epithelial cells with three different viral variants, they examined the structural details of viral attachment, entry, and budding processes using transmission electron microscopy, electron tomography, and immunofluorescence staining.
Important observations
The electron microscopy images taken 72 hours after infection revealed that most virions present in the extracellular space were attached to the epithelial cell plasma membrane. The presence of spike protein was observed on most of the virions present either in the extracellular space or within the virus-containing compartments (VCs). Interestingly, no difference in ultrastructure was noticed between cells infected with three different viral variants.
The ACE2 and TMPRSS2 were found to localize specifically at the base and surrounding of microvilli present on the cell membrane. However, the presence of these proteins was not observed in cilia. Similarly, most extracellular virions on the cell surface were found to associate with microvilli and not with cilia.
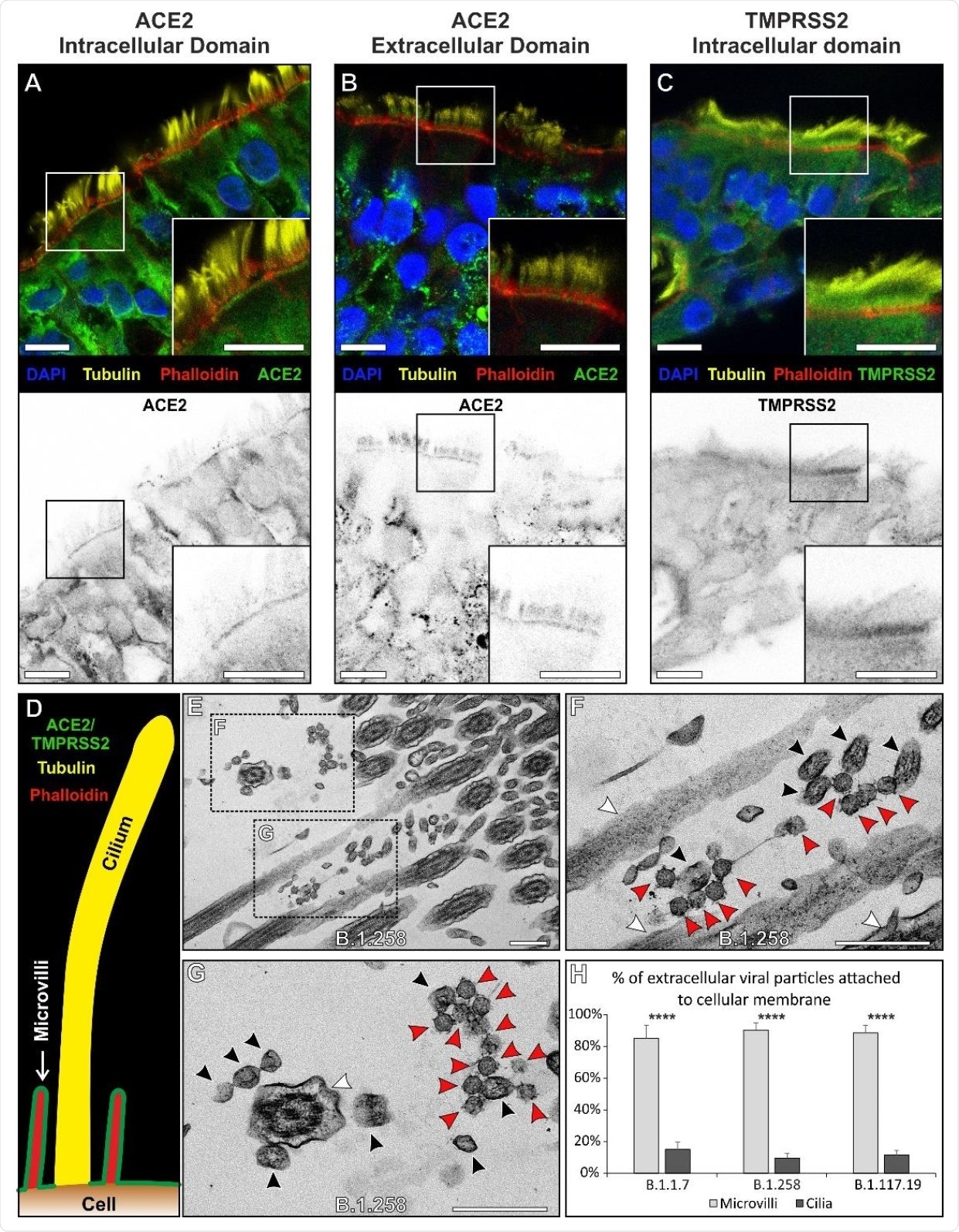
ACE2 and TMPRSS2 localises to the plasma membrane excluding cilia and this coincides with the sites of viral attachment. (A - C) Immunofluorescence antibody labelling against ACE2 and TMPRSS2 using anti-tubulin as a marker for cilia and phalloidin for the actin enriched microvilli. Both ACE2 and TMPRSS2 were found to be localised to cell surface that included the microvilli. Although there is diffuse, likely non- specific, staining everywhere with the TMPRSS2 antibody it is clearly enriched on the apical plasma membrane. Grey scale images of the staining profiles of ACE2 and TMPRSS2 show that they only extend a short difference from the cell surface unlike the tubulin staining of cilia and overlap with the phalloidin staining. (D) A diagram of the staining pattern of the antibodies. (E – G) When looking by TEM, virions (red arrowheads) are seen attached to microvilli (black arrowheads) and not to cilia (white arrowheads). (H) Quantification of the number of viral particles attached (within 3 nm) to microvilli and cilia, there is little affinity of the virus to cilia for all three isolates. Statistical significance was determine using Student’s t-tests (**** P = 0.0001). Scalebars (A – C) 10 µm and (E – G) 400 nm
Because VCs are generated as a result of successful viral entry and replication within host cells, the scientists used these compartments as a marker of cellular infection. The electron microscopy analysis identified only a few VCs in goblet cells compared to that in ciliated cells. These observations reveal that goblet cells are devoid of viral infection.
For further clarification, the scientists examined the presence of spike protein-like structures on the cell membrane as a marker of infection. The findings revealed that spike protein-like protrusions were present on the microvilli of infected ciliated cells and not on the microvilli of uninfected goblet cells. Similar protrusions were also found in some portions of the plasma membrane where virions were attached. These observations indicate two possible scenarios: 1) following viral envelope–host cell membrane fusion, the spike protein is left behind in the fused membrane patch; 2) after being expressed in the host cell, the spike protein is transported to the plasma membrane.
Regarding viral fusion, the findings revealed that the virion envelope was distended and formed a continuous connection with the host cell membrane. The nucleocapsid content of the fused virion was found to be diluted compared to whole virions present in the vicinity. This observation indicates that some nucleocapsid proteins of the fused virion have already entered the host cell.
Regarding viral budding, a neck-like structure was observed at the site of fusion, indicating that the virion is pulling away in the direction of the VC lumen. Moreover, an image of a VC with two budding virions showed a network of microtubules running into the emerging virions. Such structural arrangement indicates the transportation of viral components to the budding site.
Study significance
The study provides in-depth information about the key features of SARS-CoV-2’s life-cycle. The ultrastructural details of the virus-infected epithelial cells provided in the study could be potentially helpful in identifying therapeutic targets against COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Pinto A. 2021. Ultrastructural insight into SARS-CoV-2 attachment, entry and budding in human airway epithelium. bioRxiv. doi: https://doi.org/10.1101/2021.04.10.439279, https://www.biorxiv.org/content/10.1101/2021.04.10.439279v1
- Peer reviewed and published scientific report.
Pinto, Andreia L., Ranjit K. Rai, Jonathan C. Brown, Paul Griffin, James R. Edgar, Anand Shah, Aran Singanayagam, et al. 2022. “Ultrastructural Insight into SARS-CoV-2 Entry and Budding in Human Airway Epithelium.” Nature Communications 13 (1): 1609. https://doi.org/10.1038/s41467-022-29255-y. https://www.nature.com/articles/s41467-022-29255-y.