To date, vaccination efforts have commenced stemming the coronavirus disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Alongside vaccine campaigns, scientists race to find effective drugs to combat the infection.
Researchers at the Human Biology Division, Fred Hutchinson Cancer Research Center in Washington, United States, found that the FDA-approved, multi-kinase inhibitor Ponatinib is a potent inhibitor of the N-terminal domain (NTD) -mediated cytokine storm. Ponatinib is an oral drug used for the treatment of chronic myeloid leukemia (CML).
The drug could be used on severely ill COVID-19 patients who struggle with cytokine storms, wherein inflammatory cytokines are produced at a much higher rate than usual. The overproduction of cytokines draws the other immune cells to go to the site of injury, causing organ damage.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study background
In most patients infected with SARS-CoV-2, they experience only mild to moderate symptoms. But some people may experience severe disease, particularly the elderly and those with comorbidities.
In about 15 to 20 percent of cases, the initial infection phase is followed by a more severe event where monocytes produce a cytokine storm, rapidly releasing interleukin 6 (IL-6), Interleukin 1 beta (IL-1β), C-X-C motif chemokine ligand 10 (CXCL10), Chemokine (C-C motif) ligand 7 (CCL7), and other inflammatory molecules.
COVID-19 patients who had samples containing these molecules were more likely to have increased viral load, lung injury, loss of lung function, and a fatal clinical outcome.
Drug or drug combination that reduces the cytokine storm can help treat COVID-19. Even though the induction of the cytokine storm is regulated by IL-1 signaling, the downstream signaling pathways needed are still unknown.
Ponatinib and cytokine storm
The study, which appeared on the pre-print server bioRxiv*, showed that FDA-approved or clinical grade compounds inhibiting inflammatory cytokine production could help in curbing the pandemic.
The researchers aimed to identify drugs available for clinical use, which can help reduce the time needed to develop and distribute treatments to solve the COVID-19 pandemic.
To arrive at the study findings, the team used spontaneously immortalized monocyte-like line, THP1(3) and mammalian (HEK293) cells that generated full-length Spike subunit S1 protein, which the SARS-CoV-2 needs to enter host cells.
The team showed that 24-hour stimulation with the full-length mammalian cell-derived S1 subunit of SARS-CoV-2 spike protein led to massive upregulation of IL-1b in a dose-dependent manner.
The team also wanted to know if S1 protein could enhance the expression of cytokines in COVID-19 patients. They revealed that S1 protein stimulation leads to a marked increase in the expression of cytokines, including interleukin 8 (IL-8), IL-6, IL-1β, tumor necrosis factor (TNF), and chemokines, such as CXCL10, chemokine ligand 2 (CCL2), and chemokine ligand 7 (CCL7) in THP1 monocytes.
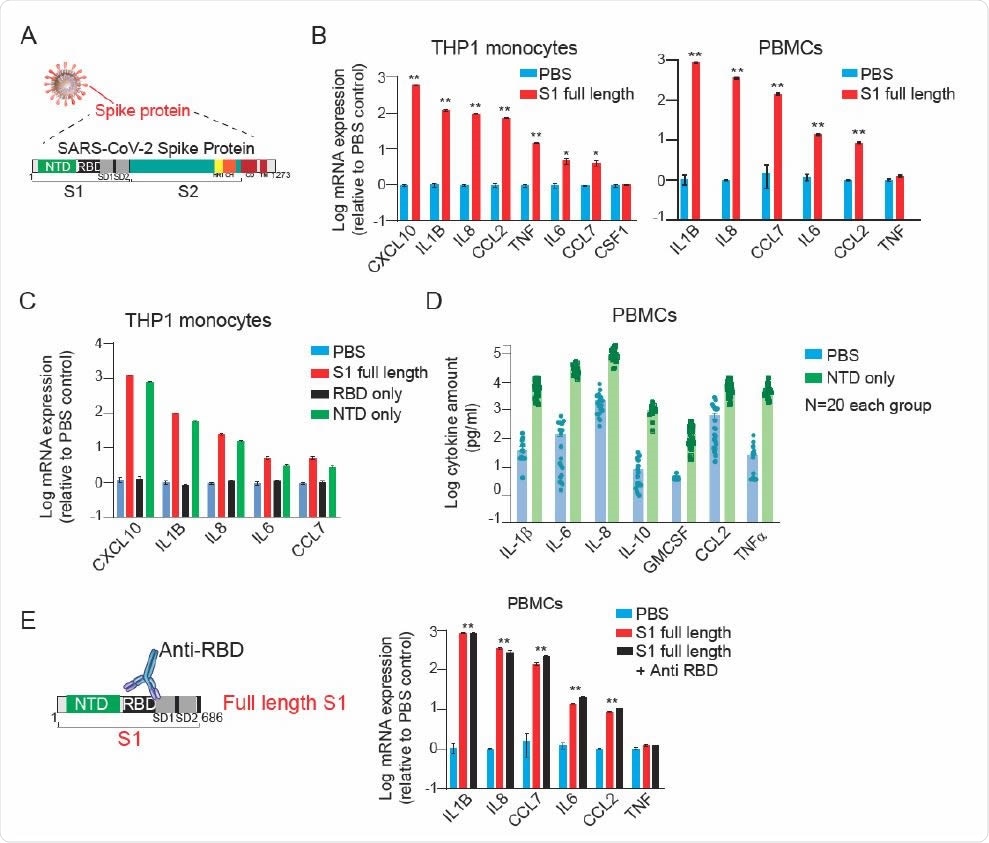
SARS-CoV-2 Spike subunit S1 protein causes a significant increase in the expression and release of a panel of cytokines in THP1 monocytes and human PBMCs. (A) A schematic showing major SARS-CoV-2 proteins and domain structure of spike protein. (B) Changes in the expression of cytokines in THP1 macrophages (left) and PBMCs (right) upon treatment with fulllength S1 subunit at 1 µg/ml for 24 hr. (C) Changes in the expression of cytokines in THP1 macrophages upon treatment with different domains of S1 subunit at 1 µg/ml for 24 hr. (D) Measurement of cytokine release from healthy donor PBMCs treated with PBS or NTD at 1 µg/ml for 24 hr. (E) Effect of an anti-RBD antibody on S1 subunit stimulated changes in the expression of cytokines in PBMCs. Gene expression was measured by qPCR. Cytokine release in the conditioned media was measured by Luminex. Full-length S1 and S1 subunits are purified from HEK293 cells.
There were similar changes in cytokines noted in healthy donor peripheral blood mononuclear cells (PBMCs) and raw mouse monocytes in response to full-length S1 protein. These findings reveal that interaction between the Spike subunit S1 protein and monocytes is enough to stimulate monocytes.
Further, the team demonstrated that the spike protein’s N-terminal domain (NTD) from the SARS-CoV-2 and emerging variants, B.1.1.7 and B.1.351, significantly induces various inflammatory molecules in monocytes and PBMCs. Moreover, they identified many protein kinases, such as JAK1, EPHA7, IRAK1, MAPK12, and MAP3K8, as vital downstream mediators of NTD-induced cytokine release.
The study also identified Ponatinib as being able to inhibit the NTD-mediated cytokine storm effectively. Overall, the team proposed that agents targeting multiple kinases may be used as a therapeutic option to treat moderate to severe COVID-19.
“More specifically, our results suggest that an FDA-approved drug, Ponatinib, could represent a strong candidate for drug repurposing efforts aimed at providing an alternative and timely treatment for COVID-19 patients exhibiting major, life-threatening symptoms,” the team wrote in the study.
There is an urgent need for effective treatments for COVID-19, particularly for those with severe disease. To date, over 137 million people have been infected with SARS-CoV-2 globally. Of these, over 2.95 million people have died.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Chan, M., Vijay, S., McElrath, J., Holland, E., and Gujral, T. (2021). Machine Learning Identifies Ponatinib as a Potent Inhibitor of SARS-CoV2-induced Cytokine Storm. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.04.07.438871v1
- Peer reviewed and published scientific report.
Chan, Marina, Siddharth Vijay, John McNevin, M Juliana McElrath, Eric C Holland, and Taranjit S Gujral. 2021. “Machine Learning Identifies Molecular Regulators and Therapeutics for Targeting SARS‐CoV2‐Induced Cytokine Release.” Molecular Systems Biology 17 (9). https://doi.org/10.15252/msb.202110426. https://www.embopress.org/doi/full/10.15252/msb.202110426.