Researchers in the United States have demonstrated how to estimate the potentially waning efficacy of vaccines designed to protect against infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The novel SARS-CoV-2 virus is the agent responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic that has now claimed the lives of more than 3 million people globally.
The team’s proposed methods address the challenges presented in assessing vaccine efficacy (VE) using blood or nasal samples that are only collected periodically during clinical trials where enrollment is staggered and crossover of placebo recipients to the vaccine arm occurs before the end of the study.
“Here, we provide valid and efficient statistical methods for estimating potentially waning VE against SARS-CoV-2 infection with blood or nasal samples under time-varying community transmission, staggered enrollment, and blinded or unblinded crossover,” writes the team from the University of North Carolina in Chapel Hill and Fred Hutch in Seattle, Washington.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The challenges faced in assessing VE
Since the novel SARS-CoV-2 was first identified in Wuhan, China in late December 2019, unprecedented progress has been made in developing vaccines and interim phase 3 clinical trial results have shown high VE against symptomatic COVID-19.
However, little is known regarding how effective the vaccines are at preventing people from developing asymptomatic infection and potentially inadvertently spreading the virus.
Most phase 3 trials have collected blood samples that can be used to identify seroconversion (the presence of anti-SARS-CoV-2 antibodies in the blood). However, economic and logistical limitations mean blood samples are only drawn infrequently, such that seroconversion is only known to occur between two timepoints that are weeks or months apart.
Such “ interval-censored” seroconversion data is more challenging to analyze than potentially “right-censored” symptomatic disease data, particularly when VE changes over time, say the researchers.
“An event time is said to be potentially right-censored if it is either observed exactly or known to be longer than the duration of follow-up,” explains Dan-Yu Lin and colleagues.
Further challenges arise when community transmission changes over time and when participants are vaccinated on different dates due to staggered enrollment or the crossover of placebo recipients to the vaccine arm before the end of the trial.
Infection is often diagnosed by reverse transcription-polymerase chain reaction (RT-PCR) testing of nasal swabs and most phase 3 trials have collected nasal swabs at participant enrollment and crossover.
However, such infrequent testing of swab samples will miss many infections since individuals may only be RT-PCR positive for SARS-CoV-2 over the course of a few days or weeks.
“Some phase 3 trials have taken nasal swabs more frequently (e.g., twice a week) on a subset of participants, and the newly launched Prevent COVID U study takes nasal swabs every day; however, frequent RT-PCR testing increases trial cost,” writes Lin and the team.
What did the researchers do?
Now, Lin and colleagues have shown how to evaluate potentially waning VE against SARS-CoV-2 infection using blood or nasal samples collected at different frequencies under the conditions of time-varying community transmission, staggered enrollment and potential blind or unblinded crossover to the vaccine arm.
The researchers demonstrated the usefulness of the statistical methods proposed by conducting extensive simulation studies that mimic the phase 3 trial of the Pfizer-BioNTech BNT162b2 vaccine and the Prevent COVID U study.
They allowed the risk of infection to vary across the calendar time and to depend on baseline risk factors, such as age, sex, race, and health conditions. They also allowed the VE against infection to depend on the time elapsed since vaccination.
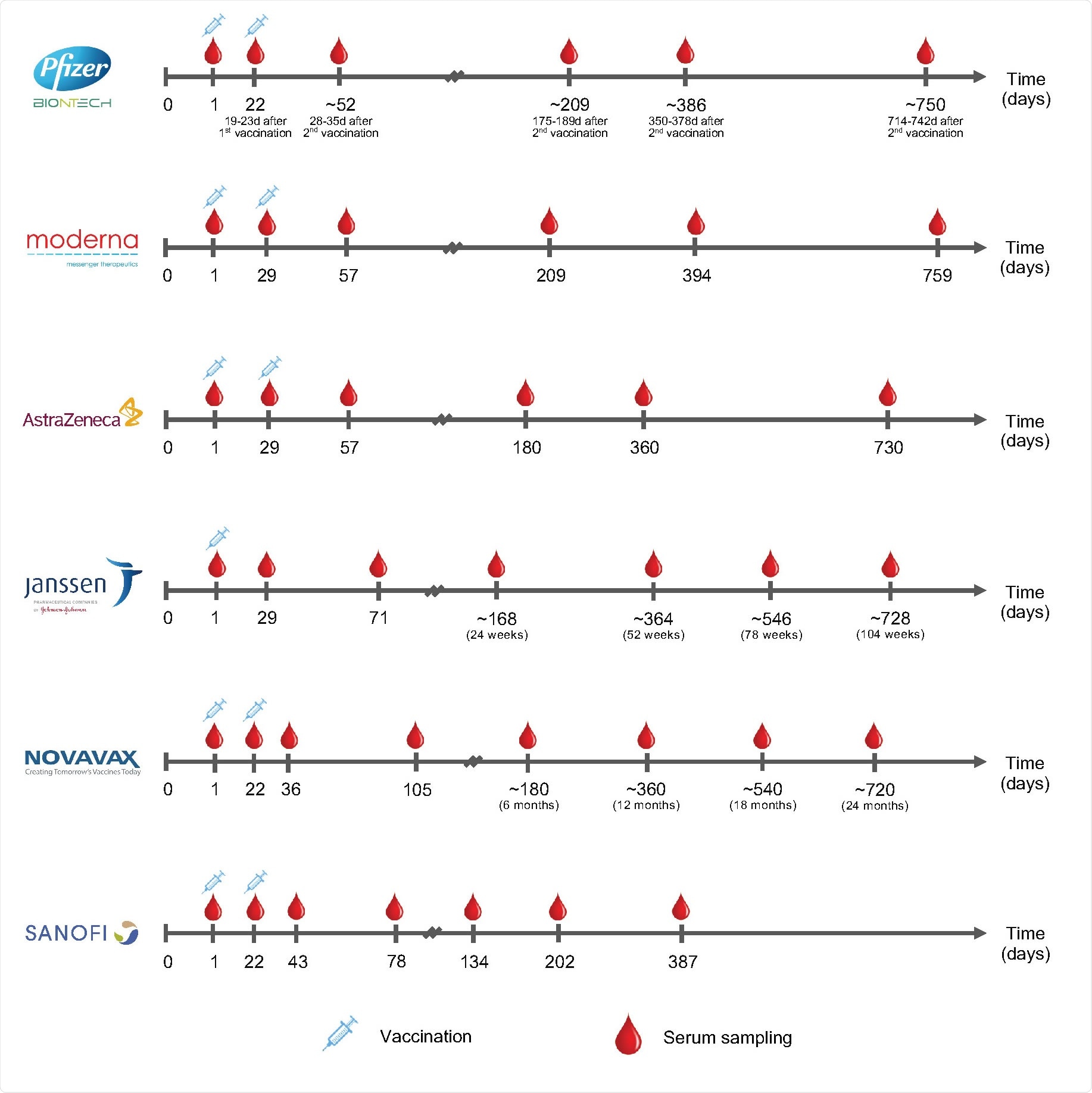
Serum sampling schedules in 6 phase 3 COVID-19 vaccine trials. The sampling time points are measured from the day of enrollment.
“The new methods provide valid and efficient estimation of three useful VE measures”
The team showed that it is possible to evaluate time-varying VE against infection using the blood samples taken during the ongoing phase 3 trials or using the nasal samples collected in studies such as Prevent COVID U.
“The new methods provide a valid and efficient estimation of three useful VE measures,” writes Lin and colleagues.
The researchers say that a significant advantage of this new approach is that it provides a unified framework for studying constant versus waning VE.
“Many studies collect both blood and nasal samples,” they write. “The proposed methods can be applied to the two types of infection data separately or as a combined endpoint, depending on the objective of the analysis and the frequency of each type of test.”
The team has implemented the proposed methods in a free software package, which is available at https://dlin.web.unc.edu/software/idove/

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources