A new study led by Hans-Martin Jäck of the Friedrich-Alexander University Erlangen-Nuremberg in Germany engineered clusters of neutralizing monoclonal antibodies using a transgenic mouse model. A unique characteristic for the antibodies was the different ways it bound to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the overall reduction in the viral spread for human ACE2.
The antibodies have the potential to work against some coronavirus variants of concern. The researchers also suggest neutralizing monoclonal antibodies could be hypothetically useful for COVID-19 therapies.
The team writes:
"We report two clusters of potent non-competing SARS-CoV-2 neutralizing antibodies providing potential candidates for therapy and prophylaxis of COVID-19. The study further supports the use of transgenic animals with human immunoglobulin gene repertoires in pandemic preparedness initiatives."
The study, "A pair of non-competing neutralizing human monoclonal antibodies protecting from disease in a SARS-CoV-2 infection model", is available as a preprint on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Immunization schedule for antibody production
The research team used the transgenic TRIANNI mouse model to create antibodies targeting the spike protein's receptor binding domain or N-terminal domain. They chose this model because of their ability to develop antibodies with a human Ig variable region and murine constant regions.
In February 2020, the research team immunized two groups of mice intramuscularly with either the SARS-CoV-2-S DNA or the SARS-CoV-2-SG-DNA. The mice were given a booster three weeks later. The mice that received SARS-CoV-2-S DNA received either the SARS-CoV-2-S DNA or the SARS-CoV-2-S protein adjuvanted with MPLA liposomes. Meanwhile, the SARS-CoV-2-SG DNA administered SARS-CoV-2-SG at week 3 or SARS-CoV-2 exosomes at week 4.
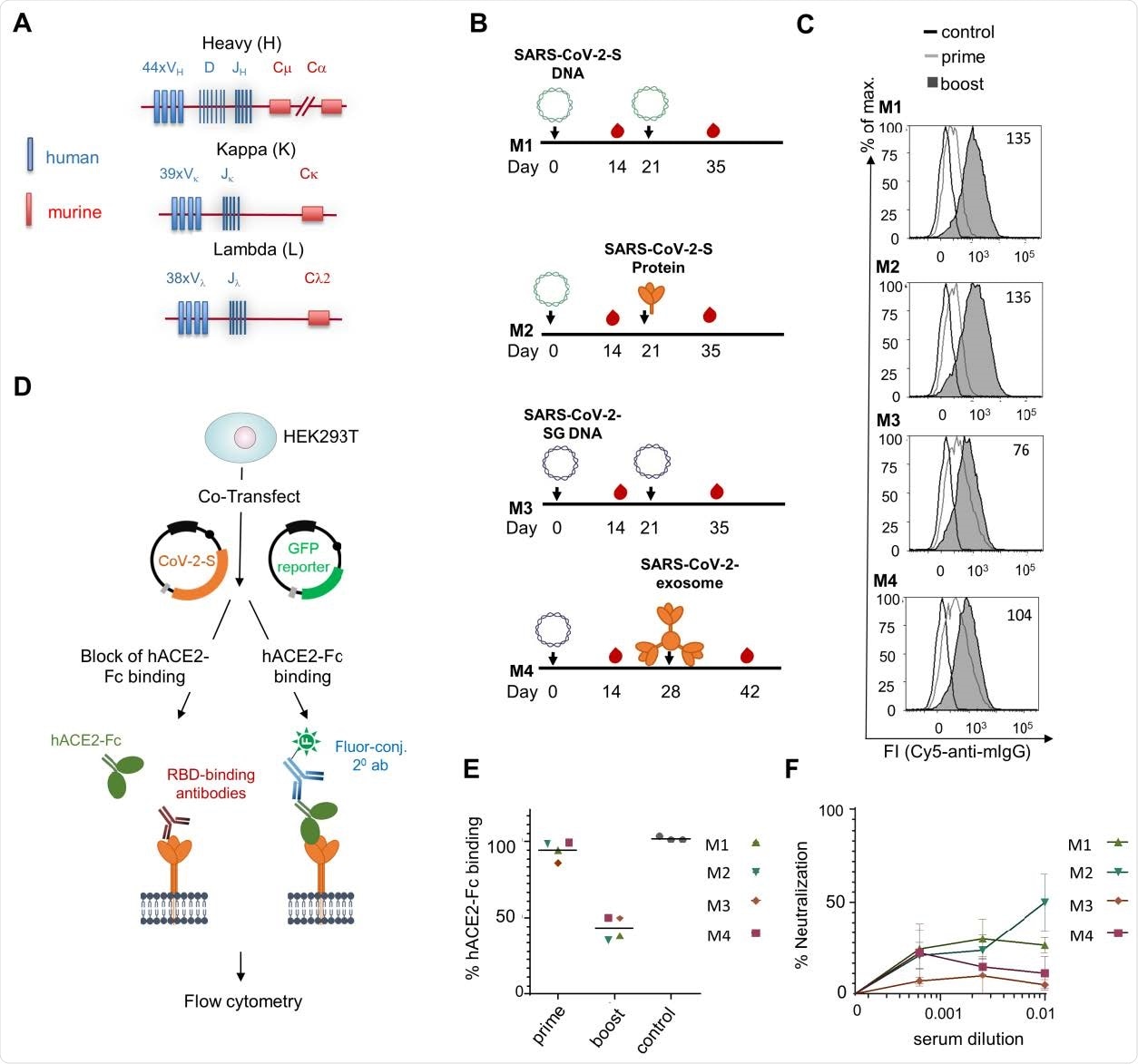
Immunization of TRIANNI mice for induction of SARS-CoV-2 neutralizing antibodies. TRIANNI mice harboring the entire human Ig variable region repertoire (A) were primed by intramuscular electroporation with expression plasmids for wild type SARS-CoV-2-S (M1, M2) or a hybrid SARS-CoV-2-S containing the intracytoplasmic domain of VSV-G (M3, M4) (B) Mice were boosted with the expression plasmids used for priming (M1, M3), soluble trimeric S protein (M2), or exosomes carrying the hybrid SARS-CoV-2-S protein (M4). (C) A flow cytometric assay assessed the binding of sera at a 1:200 dilution to the SARS-CoV-2-S protein with HEK-293T cells transiently expressing the S protein. Numbers indicate the relative mean fluorescence intensities of sera drawn two weeks after the booster immunizations. (D) Scheme of hACE2-Fc competition assay. HEK-293T cells expressing the SARS-CoV-2-S protein were incubated with hACE2-Fc fusion protein in the presence or absence of sera from immunized mice before staining with an AF647-labelled anti-human Fc antibody. (E) Competitive inhibition of hACE2-Fc binding to trimeric S protein by sera (1:200) from control mice and mice at the indicated time points after the first immunization. The mean percentage of binding as compared to control binding is shown (two experiments each performed in triplicates). (F) For the neutralization assay, Vero-E6 cells were infected with the SARS-CoV-2 isolate MUC-IMB-1 in the presence or absence of week 5 sera. SARS-CoV-2 infection was quantitated after 20 to 24 hours by staining with purified IgG from a convalescent COVID-19 patient and a fluorescence-labeled anti-human IgG using an ELISPOT reader. The mean and SEM of triplicates of one experiment are shown.
Detection and characterization of antibodies
Antibodies specific to SARS-CoV-2 were detected after the first dose, with levels further enhancing after the second dose. In total, the TRIANNI mice produced 29 hybridoma antibodies that targeted the spike protein on the cell surface of transfected HEK293T cells. Of the 29, nine neutralized SARS-CoV-2 infection at IC50 values in the subnanomolar range.
Two weeks after the immunization regimen, the researchers tested the monoclonal antibodies produced from mice by exposing them to cells transfected with SARS-CoV-2-S DNA.
The binding properties differed across monoclonal antibodies. Some bound to the receptor-binding domain and competed for ACE2 binding, others bound to the S1 subunit only, others bound to the ectodomain of the spike protein stabilized in a trimeric form and the N-terminal domain, and others showed binding to the S2 subunit only.
When testing mice serum, they found antibodies produced direct competition to the SARS-CoV-2-S protein showed reduced viral binding two weeks after the booster shot.
Researchers produced monoclonal TRES antibodies, and with a DNA prime protein boost immunization regimen, they observed mutations in the VH and VL region. The researchers suggest this means binding affinity for the antibodies can improve against SARS-CoV-2 or potentially other viruses that cause disease such as HIV.
"Transgenic animal models supporting affinity maturation of antibodies with human variable regions should be an essential part of future pandemic preparedness efforts and be explored in structure-guided vaccine antigen design against SARS-CoV-2."
The research team also tested the neutralizing power of the monoclonal antibodies on two variants of concern. One cluster of antibodies showed potential against the B.1.351 variant, but another cluster of antibodies was ineffective against both the B.1.1.7 and B.1.351 variants of concern.
While the transgenic models produced potent neutralizing antibodies, a major downfall with this method is the long time to produce them. The complete development of human antibodies took approximately 19 weeks. However, the researchers note there is potential to shorten the immunization schedule and, therefore, reduce development time by several weeks.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A pair of non-competing neutralizing human monoclonal antibodies protecting from disease in a SARS-CoV-2 infection model, Antonia Sophia Peter, Edith Roth, Sebastian R. Schulz, Kirsten Fraedrich, Tobit Steinmetz, Dominik Damm, Manuela Hauke, Elie Richel, Sandra Mueller-Schmucker, Katharina Habenicht, Valentina Eberlein, Leila Issmail, Nadja Uhlig, Simon Dolles, Eva Grüner, David Peterhoff, Sandra Ciesek, Markus Hoffmann, Stefan Pöhlmann, Paul F. McKay, Robin J. Shattock, Roman Wölfel, Ralf Wagner, Jutta Eichler, Wolfgang Schuh, Frank Neipel, Armin Ensser, Dirk Mielenz, Matthias Tenbusch, Thomas H. Winkler, Thomas Grunwald, Klaus Überla, Hans-Martin Jäck, bioRxiv, 2021.04.16.440101; doi: https://doi.org/10.1101/2021.04.16.440101 https://doi.org/10.1101/2021.04.16.440101, https://www.biorxiv.org/content/10.1101/2021.04.16.440101v1
- Peer reviewed and published scientific report.
Peter, Antonia Sophia, Edith Roth, Sebastian R. Schulz, Kirsten Fraedrich, Tobit Steinmetz, Dominik Damm, Manuela Hauke, et al. 2021. “A Pair of Noncompeting Neutralizing Human Monoclonal Antibodies Protecting from Disease in a SARS‐CoV‐2 Infection Model.” European Journal of Immunology 52 (5): 770–83. https://doi.org/10.1002/eji.202149374. https://onlinelibrary.wiley.com/doi/10.1002/eji.202149374.