As the coronavirus disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to spread worldwide, scientists race to determine vaccines and drugs that can combat the pandemic.
A team of Australian researchers at the Walter and Eliza Hall Institute of Medical Research (WEHI), the Doherty Institute, and the Kirby Institute have identified neutralizing nanobody cocktails that block the SARS-CoV-2 virus from entering cells in pre-clinical models.
The study, published in the journal Proceedings of the National Academy of Sciences of the United States of America (PNAS), identified high-affinity nanobodies against SARS-CoV-2 receptor-binding domain (RBD). These nanobody cocktails blocked the angiotensin-converting enzyme 2 (ACE2) binding with RBD variants present in human populations.
The team also found that the nanobodies can potently neutralize both the wild-type SARS-CoV-2 and the N501Y D614G variant at low concentrations.
Image Credit: Huen Structure Bio / Shutterstock
Nanobodies in medicine
Nanobodies (Nbs) are mini antibodies ten times smaller than regular antibodies produced naturally by alpacas, camels, and llamas in response to infection.
Camelid-derived single-domain antibody fragments or nanobodies offer many advantages over conventional antibodies as candidates for specific therapies. Despite being so small, they retain specificity and affinity similar to traditional antibodies while being easier to clone, express, and manipulate.
Hence, nanobodies are cost-effective and can be produced in large quantities. Critical to their use as antivirals in humans, they have also proven to be highly potent inhibitors of viral infections, highly needed amid the coronavirus pandemic.
While most nanobodies show potent neutralization against SARS-CoV-2 using in vitro assays, very few studies have examined the in vivo efficacy of nanobodies to prevent and treat COVID-19.
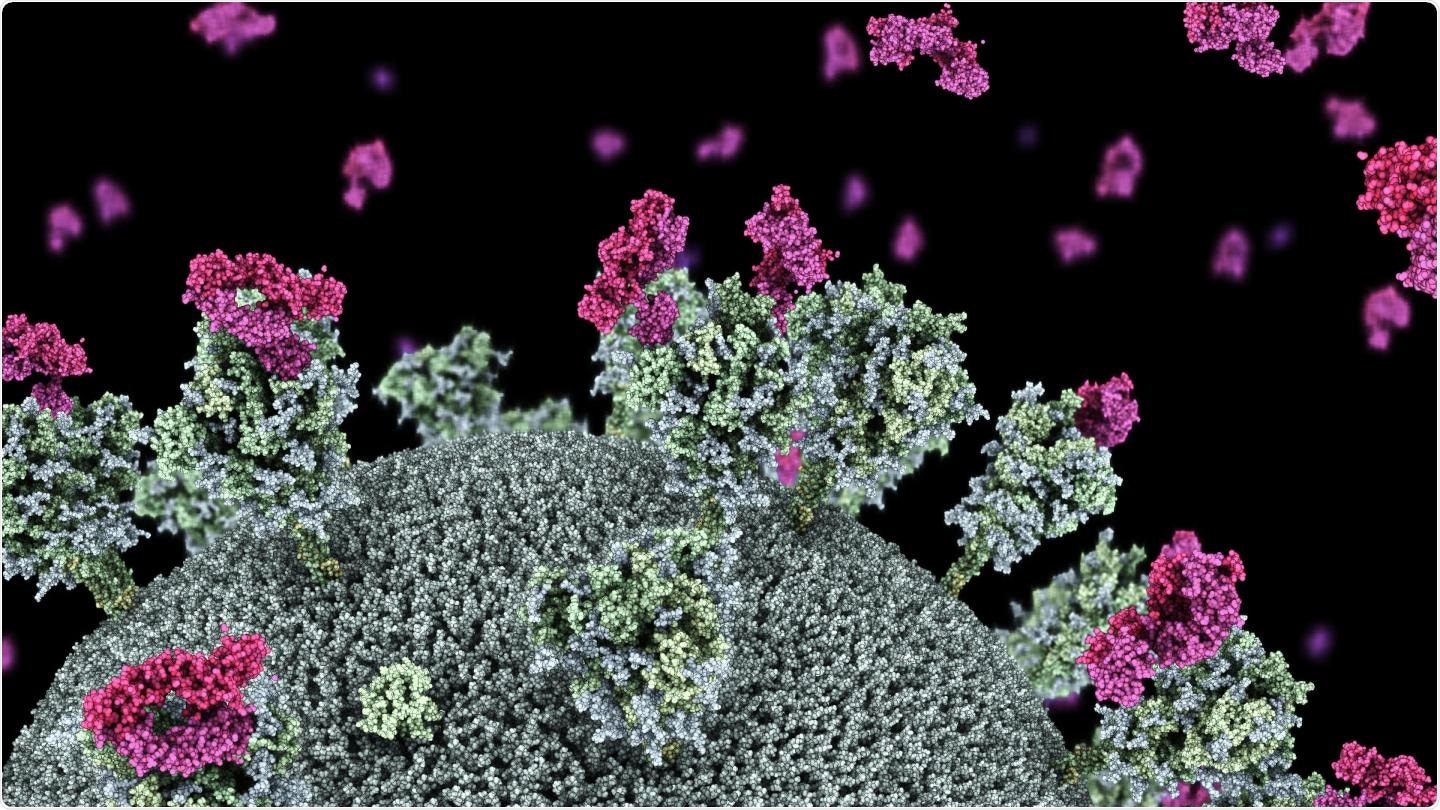
Visualization of SARS-CoV-2 virus with nanobodies (purple) attaching to the virus 'spike' protein. Image credit: Dr. Drew Berry in collaboration with Associate Professor Wai-Hong Tham
The study
The study is part of a consortium-led effort to bring together scientists in Australia to find a powerful solution to the pandemic spread, which has now caused over 147 million infections worldwide.
The team used epitope mapping, X-ray crystallography, and cryo-electron microscopy to reveal two distinct antigenic sites. The procedures also showed two nanobodies from various epitope classes attached to the spike timer.
To arrive at the study findings, the team immunized a group of alpacas with a synthetic and non-infectious SARS-CoV-2 “spike” protein to generate nanobodies against SARS-CoV-2.
The team also extracted the gene sequences encoding the nanobodies and produced millions of nanobodies in the laboratory. From there, they chose the ones that bound to the spike protein the most.
This enabled the identification of potent neutralizing nanobodies against SARS-CoV-2 and the N501Y D614G variant.
.jpg)
Cryo-EM structure of WNb 2–WNb 10–spike complex and crystal structure of WNb2-RBD complex. (A) Cryo-EM maps for spike bound to WNb 2 and WNb 10. (Middle and Right) Overall map for spike ectodomain with the best-resolved densities for six bound WNbs (three of WNb 2 and three of WNb 10) showing all RBDs in the “up” conformation. (Left) The map of an individual RBD bound to WNb 2 and WNb 10 following focused 3D classification in Region 3.1. (B) Cryo-EM structure of WNb 2 and WNb 10 bound to the RBD (Left) and with human ACE2 superimposed (Right). (C) The WNb 2 and WNb 10 binding footprint on SARS-CoV-2 RBD are highlighted as magenta and teal, respectively. The overlay of human ACE2 helices is shown in dark blue. (D and E) Interacting residues between WNb 10 and the RBD. (F) Crystal structure of WNb 2-RBD complex. (G–K) Interacting residues between WNb 2 and the RBD identified from the crystal structure. (L) Overlay of existing nanobody structures with WNb 2 and WNb 10. The codes in the brackets refer to the PDB identification codes.
Further, the team combined the two leading nanobodies into a cocktail, allowing them to test its efficacy at blocking SARS-CoV-2 from entering cells and reducing viral loads in pre-clinical models.
ANSTO’s Australian Synchroton and the Monash Ramaciotti Centre for Cryo-Electron Microscopy allowed the researchers to map how the nanobodies bound to the spike protein and how it affected the ability of the virus to bind to its human receptor, ACE2.
By mapping the nanobodies, the team identified a nanobody that recognized SARS-CoV-2, including emerging variants. It was also effective against the original severe acute respiratory syndrome coronavirus (SARS-CoV), which caused the SARS in 2002.
The study highlights the ability of the nanobodies, particularly bivalent nanobody-Fc fusions that attached to non-competing sites on the RBD, to block ACE2 receptor engagement against a panel of naturally occurring RBD variants. Also, the fusions are high affinity, with potent neutralizing activities against SARS-CoV-2.
In a nutshell, the study highlights the importance of neutralizing nanobodies against SARS-CoV-2 as an important therapeutic option. Also, nanobody and antibody mixtures that target non-overlapping epitopes on the RBD have shown promise in preventing the occurrence of resistance mutations.
“These neutralizing nanobody-Fc fusions may find an application for the passive immunization of people who are immunocompromised and may not respond as well to vaccination or for preventing outbreaks in high-risk settings, such as aged care facilities,” the researchers explained.
They added that using nanobody mixtures can be beneficial in controlling more highly infectious variants and reducing the potential for virus escape mutations to develop.