A recent astonishing feat by US researchers revealed that recombination is not only widespread in the evolutionary history of SARS-like coronaviruses, but it can also increase fitness traits of seasonal coronaviruses. Their paper is currently available on the bioRxiv* preprint server and sets the stage for phylogenetic tracking of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Since its emergence, genetic sequence data has been used to study the evolution and spread of SARS-CoV-2, a causative agent of coronavirus disease 2019 (COVID-19) pandemic. Consequently, the research community quickly embraced phylogenetic and phylodynamic methods as essential tools to study this RNA virus.
Alongside mutations caused by errors during the replication process, various recombination processes can contribute to genetic diversity in RNA viruses. Furthermore, reassortment in segmented viruses (such as rotaviruses or influenza viruses) can result in offspring that harbor segments from different parent lineages.
In any case, recombination constantly challenges phylogenetic methods, as it breaches the central assumption that the evolutionary history of individuals can be typified by branching phylogenetic trees. Consequently, recombination necessitates representation of the shared ancestry of a set of sequences as a network.
In this study, Dr. Nicola Felix Müller, Dr. Kathryn E. Kistler and Dr. Trevor Bedford from the Fred Hutchinson Cancer Research Center and the University of Washington in Seattle (US) have developed a Markov chain Monte Carlo approach to efficiently infer recombination networks from genetic sequence data under a template-switching model of recombination.
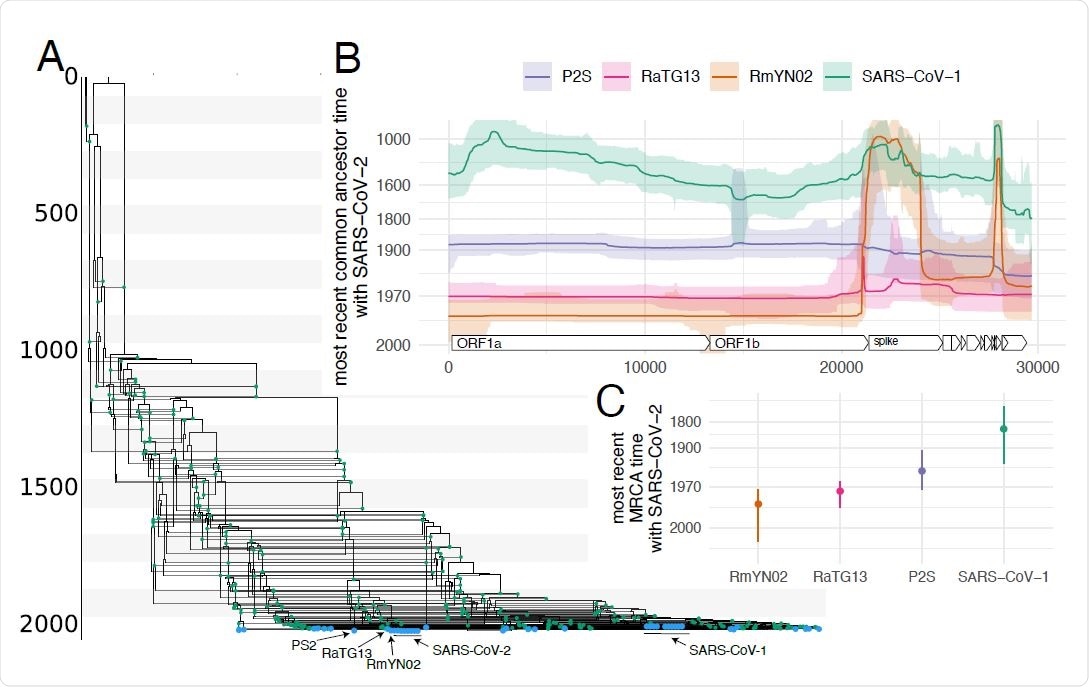
Evolutionary history of SARS-like viruses. A Maximum clade credibility network of SARS-like viruses. Blue dots denote samples and green dots recombination events. B Common ancestor times of Wuhan- Hu1 (SARS-CoV-2) with different SARS-like viruses on different positions of the genome. The y-axis denotes common ancestor times in log scale. C Most recent time anywhere on the genome that Wuhan-Hu1 shared a common ancestor with different SARS-like viruses
Assessing recombination patterns
The researchers have reconstructed the recombination history of SARS-like viruses, which includes the original SARS-CoV-1 and novel SARS-CoV-2, as well as related bat and pangolin coronaviruses. Then they have investigated in which instances different viruses last shared a common ancestor along the genome.
Recombination patterns were further appraised in the Middle East Respiratory Syndrome coronavirus (MERS-CoV) with over 2,500 confirmed cases in humans, as well as human seasonal coronaviruses OC43, 229E, and NL63 that show widespread seasonal circulation.
They next aimed to see whether recombination rates are increased on parts of the genome that also show strong signs of adaptation. This was done by computing the rates of adaption on different parts of the genomes of the seasonal human coronaviruses
In short, such an approach and this type of framework allowed combined estimation of recombination networks and rates, effective population sizes, but also parameters that delineate mutations over time from sampled genetic sequence data.
A complex evolutionary history of SARS-like coronaviruses
This paper has clearly demonstrated that recombination events are prevalent in the evolutionary history of SARS-like coronaviruses and that recombination rates in aforementioned human seasonal coronaviruses tend to vary with rates of adaptation. This implies that recombination may be beneficial to the fitness of human seasonal coronaviruses.
On the other hand, the evolutionary history of SARS-like coronaviruses was shown to be very complex and with little resemblance to tree-like evolution. In addition, the researchers have shown that recombination was only seen between closely related SARS-like viruses in the recent history of SARS-CoV-2.
One notable finding was that recombination also habitually occurs in human coronaviruses at rates comparable to reassortment in influenza viruses. Recombination breakpoints in human coronaviruses can also vary in accordance with rates of adaptation across the genomes, which suggests recombination events are positively or negatively selected based on where such breakpoints tend to occur.
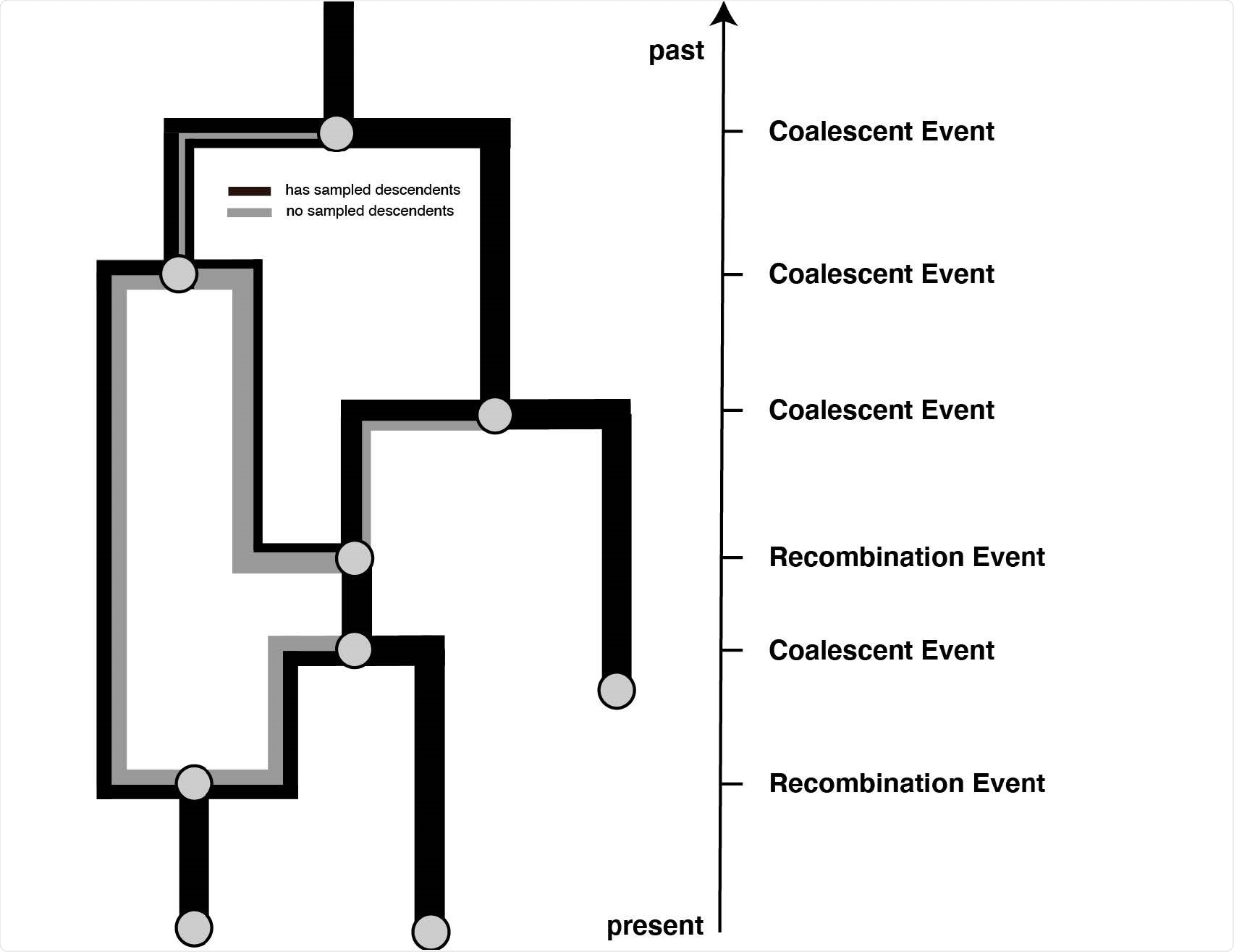
Example recombination network. Events that can occur on a recombination network as considered here. We consider events to occur from the present backward in time to the past (as is the norm when looking at coalescent processes. Lineages can be added upon sampling events, which occur at predefned points in time and are conditioned on. Recombination events split the path of a lineage in two, with everything on one side of a recombination breakpoint going in one and everything on the other side of a breakpoint going in the other direction.
Phylogenetic reconstruction
It is clear that knowing the extent to which recombination is anticipated to shape SARS-CoV-2 in the subsequent years and having the tools to identify such events and to conduct phylogenetic reconstruction will be an essential addition to our diagnostic armamentarium.
“Estimating the deleterious load of viruses before and after recombination using ancestral sequence reconstruction could help shed light on which sequences are favored during recombination,” say study authors in this bioRxiv paper.
“Additionally, having additional sequences to reconstruct recombination patterns, the seasonal coronaviruses should clarify the role recombination plays in the long-term evolution of these viruses,” they conclude.
The plausible rise of SARS-CoV-2 recombinants in the future will increase the need for methods that perform phylogenetic and phylodynamic inferences in the presence of recombination. Finally, the applicability of recombination models will open the door for the more pervasive use of models for studying the spread of pathogens.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources