The current coronavirus disease 2019 (COVID-19) pandemic has already claimed over three million lives globally. However, this is only a minute fraction of the number of infections. It is difficult to predict which patients will develop severe or critical disease, since they make up a small but sizable minority of total infections.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
The importance of cellular immunity
Following recovery from COVID-19, immunity sets in, but its extent and duration are still to be determined. Vaccination has also been initiated on a war footing in some countries, and has been demonstrated to elicit a strong immune response.
However, as new and potentially evasive mutants continue to emerge, and as India faces an unimaginable COVID-19 crisis, it is obvious that the virus is far from diminishing as a worldwide threat to life, health and economic wellbeing. Antibody-mediated protection has been extensively, though incompletely, studied, but cellular immunity is key to providing infected cells and clearing the infection, even without seroconversion.
The current paper aims to understand how cellular immunity impacts protection against reinfections.
HLA-TCR interplay
Both CD4+ and CD8+ T cells have been found to recognize specific SARS-CoV-2 epitopes, including both structural and non-structural proteins. Some of these cells have been found in naïve individuals, too, perhaps due to cross-reactive endemic coronaviruses that circulate widely.
Two factors determine the range of epitope recognition in SARS-CoV-2 infection. One is the T cell repertoire (TCR), the unique sequence of gene segments that allows a TCR to recognize one and only one antigen, and the other is the human leukocyte antigen (HLA) genes.
In mammals, cells express up to six distinct HLA class I alleles, which determine how and which antigens are presented to the immune system. The type of allele influences both the individual’s susceptibility to viral infection and the outcome.
Some large genome-wide association studies (GWAS) have failed to show clear correlations between the HLA alleles and the incidence or severity of COVID-19. However, since the HLA alleles determine antigen presentation, they work together with the selection of TCR clones to shape the T cell repertoire following infection.
This, in turn, plays an important role in the immune response to the infection, including both the response of cytotoxic T cells in clearing both the infected cells and viral particles from the body, and memory cells that preserve immunological memory to ensure a quick and specific response in case the pathogen is re-encountered.
Public TCR responses
This virus appears to elicit public T cell responses, which means that the complementarity-determining region (CDR) sequences of the epitope-recognition regions remain the same in different TCRs within individuals and across different people. This is despite the large potential repertoire of 1015 TCRs.
Study details
To understand this better, the researchers used a novel technique to correlate T cell specificity, HLA variation, conserved regions in pairs of both α/β TCR repertoires, and the CD8* T cell phenotype when exposed to this virus.
They examined these parameters in over 100 million CD8+ T cells from 76 individuals. These people came from acutely infected, convalescent, or infection-naïve groups.
They found almost 650 epitopes spanning the various viral proteins, which were presented by four HLA alleles. These are mostly spared by the currently circulating variants of concern (VOCs), indicating that they can be targeted by T cells to prevent infection following vaccination or recovery.
In fact, though 85 epitopes were present in the spike protein variant that is used in most current vaccines, the VOCs affected only six of them.
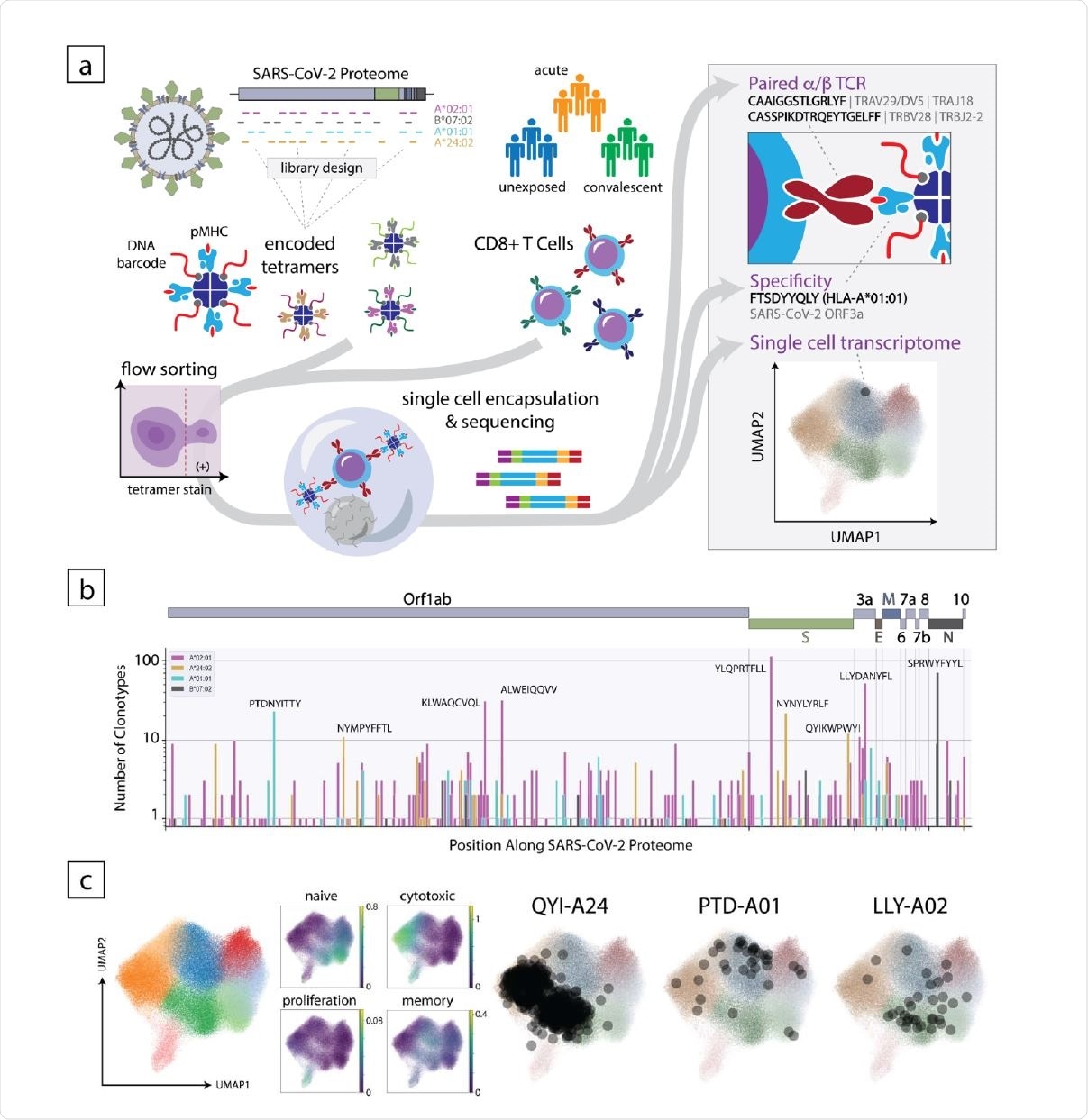
Overview of the experimental approach to decode the CD8+ T cell response to SARS-CoV-2. (a) Schematic of the method where encoded 5 tetramer libraries, designed independently for each HLA allele to span the entire SARS-CoV-2-proteome, are used to stain enriched CD8+ cells from subject PBMCs, which are then sorted and subjected to singlecell sequencing (left). Using this approach, TCR sequence, peptide/HLA specificity and transcriptomic features are simultaneously acquired for each cell (right). (b) Clonotype specificity detected by HLA allele and epitope across the SARS-CoV-2 proteome. A scheme of the viral ORF structure is shown at the top. Bar colors denote HLA allele. Amino acid sequences of epitopes recognized by the largest number of T cell clonotypes are shown next to the corresponding bar (c). Single-cell transcriptomic analysis showing global UMAP clustering, scoring by functional gene set, and projections onto the transcriptomic UMAP for T cells with specificity toward select epitopes in convalescent individuals. QYI-A24, PTD-A01, and LLYA02 correspond to QYIKWPWYI in A*24:02, PTDNYITTY in A*01:01, and LLYDANYFL in A*02:01, respectively
Changes with clinical course
The researchers found that the frequency of SARS-CoV-2-specific T cells to the top five epitopes related to each of the four HLA variants was greatly reduced during acute infection compared to convalescent patients.
Memory T cells induced by prior influenza and Epstein-Barr virus were also reduced but not cytomegalovirus.
This is as expected, given the lymphopenia in acute COVID-19 patients. in addition, COVID-19 induces a shift of antiviral T cells between different pools, perhaps in response to exposure history and also determined by epitope specificity.
With HLA B*07, in naïve subjects, the top three epitopes were concentrated in the central memory pool, while in convalescents, the reactivities were broadly distributed across central memory and terminal effector clusters of many epitope-specific T cells.
Further analysis shows that these cells have a robust memory phenotype as well as cross-reactive TCR responses in the naïve subjects.
Response to SARS-CoV-2 shows HLA-dependence
Secondly, T cells specific for SARS-CoV-2 showed varying frequencies in naïve individuals, depending on the HLA allele. For instance, HLA-A*02, A*24, and A*01-restricted dominant epitopes showed high T cell frequencies in over 40% of convalescents, but less in naïve individuals.
This was not seen with HLA B*07:02, with the strongest epitopes showing uniform frequencies across all individuals, exposed or not. Reactive CD8+ T cells to SPR-B07 occurred in almost 80% of naïve individuals and mostly linked to the central memory cell pool, indicating that it would be immunodominant in case of infection. Indeed, reactivity was noted in 100% of convalescent subjects, agreeing with the earlier findings of early memory formation in this infection.
The findings indicate that, unexpectedly, the T cells specific to these epitopes showed a diverse repertoire, dominated by pre-existing polyclonal immunity elicited by HLA-B*07:02-restricted TCRs reactive to multiple SARS-CoV-2 epitopes. The least public of the epitope-specific repertoires thus provided the largest pool of pre-existing cell immunity.
Apparently, the latter is competent to present nucleocapsid epitopes homologous between SARS-CoV-2 and the endemic human coronaviruses.
The responses to HLA A*02-, A*24-, and A*01-restricted epitopes were more often associated with “public” CDR3 motifs (shared between individuals), always showing the use of the variable (V) gene segment in the α− and/or β− chains.
A*02 showed insignificant activation with HCoV spike epitopes but marked activation with B*02-bound epitopes. The immunodominant responses in A*02 individuals were mostly due to the induction of T cells newly primed by specific SARS-CoV-2 epitopes.
Effective concentrations show similarity
Moreover, three B*07-reactive TCRs from COVID-19 patients showed almost identical 50% effective concentrations (EC50) for SARS-CoV-2 and HCoV epitopes.
While two TCRs from naïve subjects showed similar EC50s for both types of epitopes, the EC50 of the third was ten-fold higher for the HCoV epitope. Thus, homologous epitopes are reflected in functional cross-reactivity due to pre-existing cellular immunity to HCoVs.
What are the conclusions?
The findings show that HLA variation plays an important role in shaping the diversity of CD8+ T cell repertoires in response to exposure to SARS-CoV-2. The phenotype and the formation of memory cells may be partly, at least, dependent on the presentation of specific HLA-restricted epitopes.
While pre-existing memory CD8+ T cells reactive to SPR-B07 were found in the large majority of naïve subjects and in all convalescents, the results do not provide evidence for T cell exhaustion. However, this study examined only blood cells which may not always reflect events in the lung.
Dampening of all specific CD8+ T cells is also seen in this study, not only for cross-reactive T cells primed by endemic HCoVs but for newly primed SARS-CoV-2-specific cells. Strangely, this occurs precisely when their role is most important.
The same paucity is observed for cells reactive to influenza and EBV in patients with acute COVID-19, hinting at a failure of adaptive cellular immunity. “One might proposeSARS-CoV-2’s noxiousness stems from a broad obstruction of antiviral CD8+ T cell responses.” This might be, the researchers suggest, due to T regulatory (Treg) cell overactivity, which suppresses CD8+ T cells in severe COVID-19.
Our data suggest a strong association between HLA genotype and the CD8+ T cell response to SARS-CoV-2, which may have important implications for understanding herd immunity and elements of vaccine design that are likely to confer long-term immunity to protect against SARS-CoV-2 variants and related viral pathogens.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.