As the coronavirus disease (COVID-19) pandemic continues to spread globally, scientists are still learning about essential viral biological features of its causative pathogen, the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Answers to these questions are essential to forming a general understanding of viral pathogenesis, which helps us develop effective prevention and treatment strategies for COVID-19.
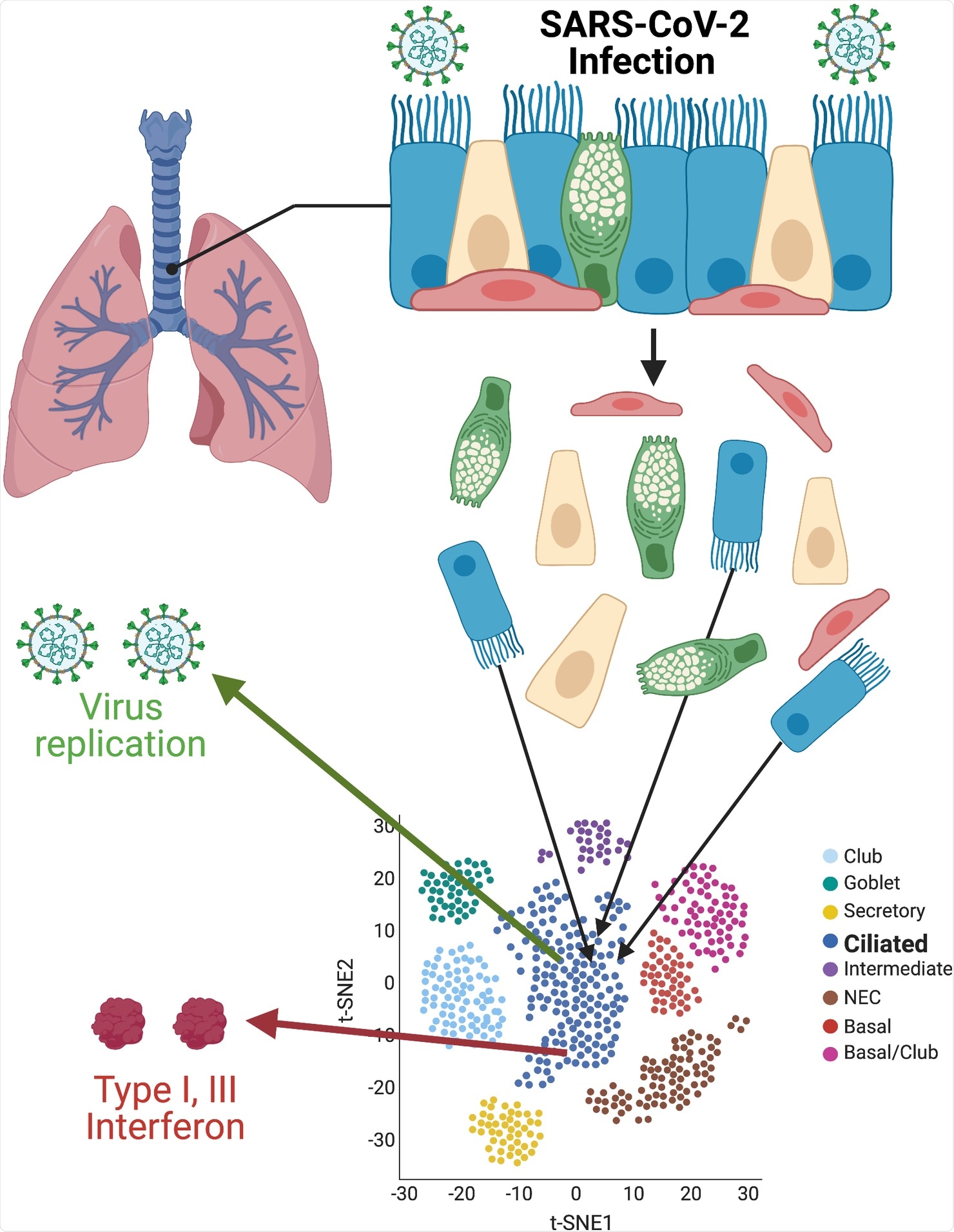
Single-cell resolution of SARS-CoV-2 infection. Primary cultures of well-differentiated bronchial epithelial cells, grown on permeable supports at ALIs, are infected with SARS-CoV-2, then separated for subsequent single-cell RNA sequencing. Using t-SNE statistical methods, cells with similar transcriptome profiles are group-clustered on two-dimensional plots. Individual cells containing SARS-CoV-2 RNAs and antiviral interferon transcripts are identified within clusters. Images were created with BioRender.com. ALI, air–liquid interface; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; t-SNE, t-distributed stochastic neighbor embedding.
Viral entry
SAR-CoV-2 binds to the human angiotensin-converting enzyme 2 (ACE2) receptor on the surface of cells lining the respiratory tract. The virus then travels through the tract, a complex system, encompassing nasal passages, large and small airways, bronchioles, and alveoli, where gas exchange occurs.
The complex relationships between initial infection and the succeeding coordinated antiviral responses occur in the respiratory epithelium, determining clinical outcomes.
The sensitive assays that detect viral RNA and protein in COVID-19 patient samples showed sino-nasopharyngeal, airway, and alveolar cell infection. In animal studies, researchers determined the accumulation and distribution of SARS-CoV-2 in the respiratory system.
Human nasal, airway and alveolar epithelial cell cultures have shed light on the complexity of the respiratory system. Studying these cells can help determine how the virus spreads in the body and the mechanisms utilized for its spread.
Early SARS-CoV-2 infection
U.S.-based researchers – at Loyola University Chicago and the University of Iowa – extended this research but focused on the high-resolution images of the first stages of SARS-CoV-2 infection and the innate immune responses that followed.
The team used infection of human bronchial epithelial-derived air–liquid interface (ALI) cultures and single-cell RNA sequencing to arrive at the study findings. The research team focused on virus tropism and validated previous reports that infection is initially localized in ciliated cells.
Motile cilia or moving cilia are found in the lungs, respiratory tract, and middle ear. The cilia perform waving or beating motion to keep the airways clear of mucus, dirt, or pathogens, allowing for easy breathing. Impaired ciliary movement in the respiratory tract may impede the clearance of harmful pathogens and foreign bodies inhaled.
Further, the researchers found that one of the virus-activating proteases, transmembrane protease serine 2 (TMPRSS2), was highly expressed in ciliated cells. Hence, in the ALI system, the TMPRSS2, is central to tropism as the primary ACE2 receptors.
When infection progresses, the virus may spread even to other cells, including the basal and club cells. Basal cells are crucial forerunners for all surface epithelial cell types. These cells contain regenerative capacity, which can be affected by SARS-CoV-2 infection.
Interferon and cytokines
The researchers also explored the relationships between SARS-CoV-2 RNA accumulation and antiviral interferon transcription. The single-cell resolution showed that only a tiny part of strongly infected ciliated cells expressed interferons.
However, these cells produced enough type I and III interferons, stimulating a pan-ALI expression of interferon-stimulated genes (ISGs). Suppose there is a sufficient amount of interferons early in the infection. In that case, it can help reduce disease in animal models of both SARS-CoV-2 and the closely related betacoronavirus severe acute respiratory syndrome coronavirus (SARS-CoV).
Apart from interferons, during COVID-19, the airway cells release chemokines such as CXCL10 and cytokines such as interleukin (IL)-6 and IL-8, which trigger the recruitment of adaptive immune cells and inflammation.
However, in some people, they develop a hyper-inflammatory response, which is a clinical feature in severe COVID-19.
Therapeutics for COVID-19
The studies noted that antiviral therapeutics should stem overactive inflammation while preserving antiviral interferon-stimulated effectors. They added that such manipulation is possible since infected ALI cultures exposed to the SARS-CoV-2 RNA replication inhibitor, Remdesevir, expressed ISGs but not IL-6.
These findings call attention to dosing and timing regimens for direct-acting antiviral drugs so that infections are suppressed while preserving controlled, appropriate immune responses,” the researchers explained.
The studies demonstrate the utility of ex vivo infection models in exploring the effects of SARS-CoV-2 in the body, which has now infected over 152.88 million people and killed more than 3.20 million people.
Further, these studies show the mechanism on how the virus enters and infects the body in the early stages of exposure.