Researchers in the United States have developed a novel pipeline for high-throughput, automated annotation of the genes, proteins, and functional domains in the genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The team from IBM Almaden Research Center in San Jose, California, says the tool offers the advantage of not having to rely on the use of a single reference genome, which can present limitations as the virus continues to evolve new variants.
Since the COVID-19 outbreak first began in Wuhan, China, in late December 2019, intense research efforts have been made globally to sequence the SARS-CoV-2 genomes observed in infected patients with near real-time efficiency.
"In order to capitalize on this large and growing corpus of data, high throughput computational methods must be developed for rapid, high accuracy analysis to deliver the molecular targets that are actually under evaluation for drug development, vaccine specificity, and diagnostic testing," says the team.
Now, Kristen Beck and colleagues have developed a novel annotation pipeline that generated gene, protein, and domain data across 66,905 publicly available SARS-CoV-2 sequences.
The data provide molecular targets efficiently and accurately across the entire SARS-CoV-2 proteome and all genomes analyzed.
A pre-print version of the research paper is available on the bioRxiv server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Tools that rely on a single reference sequence are limited
Commonly referred to as the "Wuhan reference genome," the first sequenced SARS-CoV-2 genome was published in January 2020 and quickly became the accepted reference standard.
However, since then, tens of thousands of SARS-CoV-2 genomes have been published every week.
Several viral genome annotation methods such as VAPiD, Prokka, and InterProScan are available that aim to provide autonomous (no reference genome required) annotation of genes and proteins.
"Yet, many of these tools do not provide sufficient accuracy with 'off the shelf' use and have not yet been applied at scale as the available SARS-CoV-2 sequence data grows," say the researchers.
Furthermore, several SARS-CoV-2 variants have emerged, including the D614G variant, which arose earlier in the pandemic, and the more recently emerged B.1.1.7 variant that now accounts for the majority of new cases in the United States. The B.1.17 variant contains an N501Y mutation that enhances the binding of the viral spike protein to host cell receptors.
The mutations that occur in these variants can present challenges in applying the autonomous genome annotation method.
As an alternative method, alignment to the Wuhan reference genome can be made using tools such as NextStrain's Augur, or the UCSC SARS-CoV-2 genome browser.
This type of "supervised" analysis employs published gene data to extract sequences from the genome of interest based on positional and sequence similarity to a reference genome.
However, a reference-dependent approach presents limitations as the evolving SARS-CoV-2 is currently estimated to mutate approximately twice a month.
What did the researchers do?
The researchers used a combination of state-of-the-art tools and custom calibration tools to develop a semi-supervised genome annotation pipeline. They applied this method to 66,905 SARS-CoV-2 genomes to identify the gene, protein, and functional domain sequences within each genome.
The team identified a comprehensive set of known proteins with an average set membership accuracy of 98.5%
"We were able to achieve complete or near-complete protein set membership for all genomes," says Beck and colleagues. "Each protein is a translated gene sequence, and thus the equivalent gene identification accuracy is also achieved."
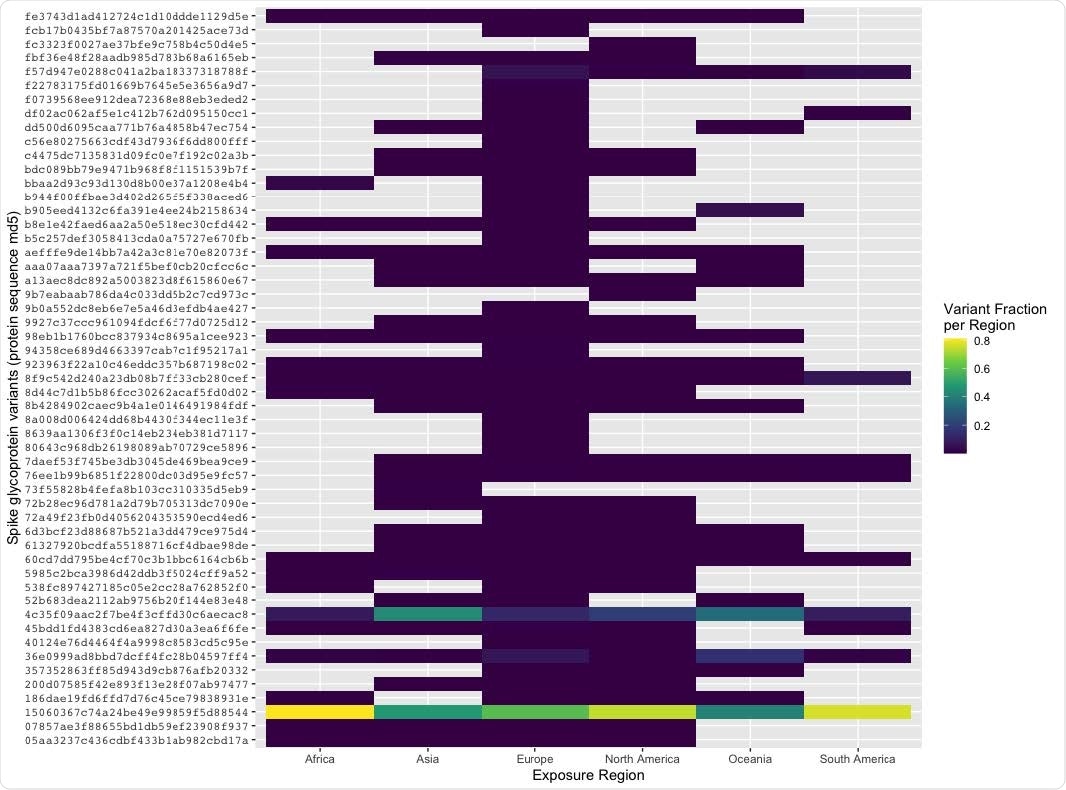
Spike glycoprotein variants observed in SARS-CoV-2 genomes over time and geography. Each line represents the cumulative frequency per variant (orange: D614G, green: UniProt ID P0DTC2, olive: P1140X, pink: S2 cleavage product) in 6a. Low frequency S protein sequences (<5 observations) are removed from plotting for simplicity. In 6b, the proportion of spike glycoprotein variants differ by exposure region. Proportion is calculated per variant to allow inter-region comparisons.
How did the method compare to other tools?
Compared with other published tools such as Prokka and VAPiD, the approach identified 6.4 and 1.8 times more protein annotations, respectively.
The method yielded almost 13 million new molecular target sequences that can be accessed through the IBM Functional Genomics Platform – a tool made freely available to the global research community.
Some of the sequences identified were conserved across time and geographical location, while others represented emerging variants.
Furthermore, for spike protein domains, the team achieved a greater than 97.9% sequence identity to references and identified variants of the spike receptor-binding domain.
"Our pipeline correctly identified key D614G and N501Y variants that have been previously observed and experimentally validated, further indicating its accuracy," writes the team.
The method could be used to inform vaccine specificity
"Here, we present a novel semi-supervised pipeline to annotate gene, protein, and functional domain molecular targets from SARS-CoV-2 genomes and demonstrate the resulting accuracy against known reference data and other bioinformatics tools," say the researchers.
Beck and colleagues say that as vaccination rollout continues during the ongoing pandemic, this method could be used to efficiently monitor and track emerging protein variants to inform vaccine specificity and host protein binding affinity.
"Additionally, as future work, further confirming the in silico predicted sequences using a structural model will allow for refinement of the protein sequences and key domains to expand our understanding of interaction with host proteins, antivirals, or diagnostics," concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Beck K, et al. Semi-supervised identification of SARS-CoV-2 molecular targets. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.05.03.440524, https://www.biorxiv.org/content/10.1101/2021.05.03.440524v1
- Peer reviewed and published scientific report.
Beck, Kristen L., Edward Seabolt, Akshay Agarwal, Gowri Nayar, Simone Bianco, Harsha Krishnareddy, Timothy A. Ngo, Mark Kunitomi, Vandana Mukherjee, and James H. Kaufman. 2021. “Semi-Supervised Pipeline for Autonomous Annotation of SARS-CoV-2 Genomes.” Viruses 13 (12): 2426. https://doi.org/10.3390/v13122426. https://www.mdpi.com/1999-4915/13/12/2426.