In SARS-CoV-2, the S1 subunit of the spike protein helps the virus attach itself to the host receptors, and the S2 subunit is involved in membrane fusion. Most antibodies target the S1 subunit, which is under selective pressure, leading to the emergence of new variants. The S2 subunit is more conserved, however, likely because of low immune pressure and the need to maintain its functionality.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Previous reports have indicated that antibodies binding to the S2 subunit could potentially neutralize distantly related coronaviruses such as influenza and HIV-1. Developing broadly neutralizing antibodies that target the conserved spike protein regions could help develop new vaccines that could protect against current and new variants.
In a paper published on the bioRxiv* preprint server, researchers report new monoclonal antibodies that target the S2 subunit and that have broad neutralization activity against several betacoronaviruses.
Characterizing antibodies
To identify such monoclonal antibodies, the team investigated IgG+ memory B cells from three COVID-19 convalescent sera. They identified five monoclonal antibodies that bind to spike protein coronaviruses that infect humans, sarbecoviruses, merbecoviruses, and embecoviruses.
Of these five, the team selected one named S2P6 for further investigations. This antibody had the highest preference for SARS-CoV-2 and SARS-CoV spike protein, followed by MERS-CoV, and OC43. This shows the potent cross-reactivity of the monoclonal antibody to human-infecting coronaviruses.
The antibody completely prevented infection of Vero-E6 cells expressing TMPRSS2, but not cells without TMPRSS2. It was also able to neutralize pseudotyped viruses of the different variants of concern of SARS-CoV-2 and other coronaviruses like SARS-CoV, MERS-CoV, and OC43.
Using peptide mapping, the team found that all five identified monoclonal antibodies bind to peptides located in the stem helix in the S2 subunit. Using cryo-electron microscopy of the spike protein complex with the antibodies, the researchers confirmed they bind to the spike protein stem helix structure, likely disrupting its quaternary structure.
Less than about 0.06% of the SARS-CoV-2 genome sequences reported so far have any mutations in the spike protein residues between 1146 and 1159, the region where S2P6 binds. None of the current variants of concern have any mutations in this region.
Experiments suggested the S2P6 antibody blocks cell-cell fusion, and is likely the main mechanism for its neutralization activity. Further crystal structure analysis revealed the key residues on the spike protein responsible for binding to the S2P6 antibody.
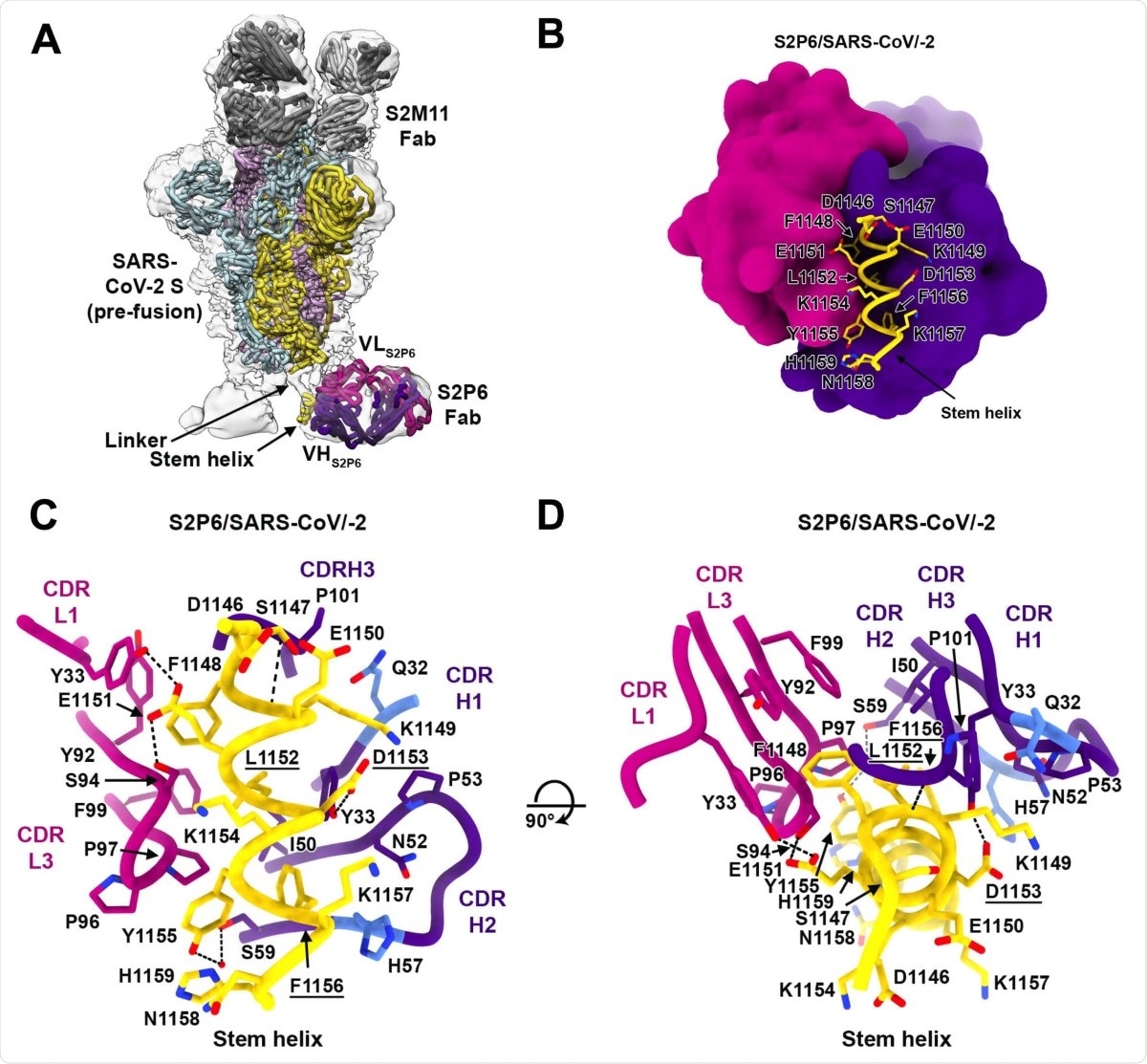
Structural basis for the broad S2P6 cross-reactivity with a conserved coronavirus. stem helix peptide. (A) Composite model of the S2P6-bound SARS-CoV-2 S cryoEM structure and of the S2P6-bound stem helix peptide crystal structure docked in the cryoEM map (transparent gray surface). SARS-CoV-2 S protomers are colored pink, cyan and gold, the S2P6 Fab heavy and light chains are colored purple and magenta and the S2M11 Fab heavy and light chains are colored dark and light gray, respectively. (B) Ribbon diagram of the S2P6 Fab (surface rendering) in complex with the SARS-CoV-2 S stem helix peptide (yellow ribbon with side chains rendered as sticks and labeled). (C-D) Ribbon diagram in two orthogonal orientations of the S2P6 Fab bound to the SARS-CoV-2 S stem helix peptide showing a conserved network of interactions. Only key interface residues and the S2P6 CDR loops are shown for clarity. Residues Q32 and H57, that are mutated during affinity maturation of the S2P6 heavy chain, are colored blue. Hydrogen bonds are indicated with dashed lines. Residues substituted in the escape mutants isolated are underlined.
The team also performed passages of the chimeric virus with SARS-CoV-2 spike protein in the presence of S2P6 to determine viral escape mutants. They found after two passages, the antibody neutralization was gone, and five different resistant mutations emerged. These mutations have not been seen much in the circulating SARS-CoV-2 strains.
Broad neutralizing capability
The authors next tested the effect of the antibody in treating Syrian hamsters infected with SARS-CoV-2, wild-type and the B.1.351 variant. The S2P6 antibody reduced viral loads in the lungs on the animals considerably for both the variants.
Tests revealed the S2P6 antibody likely arose in response to OC43 infection, and by natural infection with SARS-CoV-2 or HKU1 coronaviruses, it mutated to target these viruses also. The antibodies identified here likely arose from HKU1 infection and later became cross-reactive because of mutations.
Antibodies targeting the stem-helix of the spike protein were found at a low frequency in people who had recovered from COVID-19 and those vaccinated with the mRNA vaccine, suggesting production of such antibodies is relatively rare.
Because of the low levels of antibodies targeting the spike protein stem helix in convalescent patients or vaccinated individuals, it will be challenging to develop vaccines that can broadly target betacoronaviruses. Such efforts may require using advances such as computational protein design and vaccine design approaches focused on epitopes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Pinto, D. et al. (2021) A human antibody that broadly neutralizes betacoronaviruses protects against SARS-CoV-2 by blocking the fusion machinery. bioRxiv. https://doi.org/10.1101/2021.05.09.442808, https://www.biorxiv.org/content/10.1101/2021.05.09.442808v1
- Peer reviewed and published scientific report.
Pinto, Dora, Maximilian M. Sauer, Nadine Czudnochowski, Jun Siong Low, M. Alejandra Tortorici, Michael P. Housley, Julia Noack, et al. 2021. “Broad Betacoronavirus Neutralization by a Stem Helix–Specific Human Antibody.” Science 373 (6559): 1109–16. https://doi.org/10.1126/science.abj3321. https://www.science.org/doi/10.1126/science.abj3321.