Coronavirus disease 2019 (COVID-19) is a viral infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This disease caused the most severe pandemic of this century, with almost 150 million documented infections, over three million deaths, and is still devastating many countries with successive waves of infection.
In the absence of any specific and effective antiviral, the primary intervention that may bring an end to the pandemic is vaccination. More than a score of newly developed vaccines against COVID-19 have been rolled out, all over the world, following emergency use authorization. Tens of millions of doses have been administered.
However, as newer and more infectious, and possibly more virulent, variants of the virus emerge, and as vaccination delays are becoming obvious in large segments of the population globally, it is clear that the pandemic is far from over. Therefore, the need for antivirals remains as urgent as ever.
A new paper in the Journal of Molecular Modeling reports the discovery of promising antiviral hit compounds through the use of computational modeling studies on natural compounds with inhibitory activity against one or more viral enzymes.
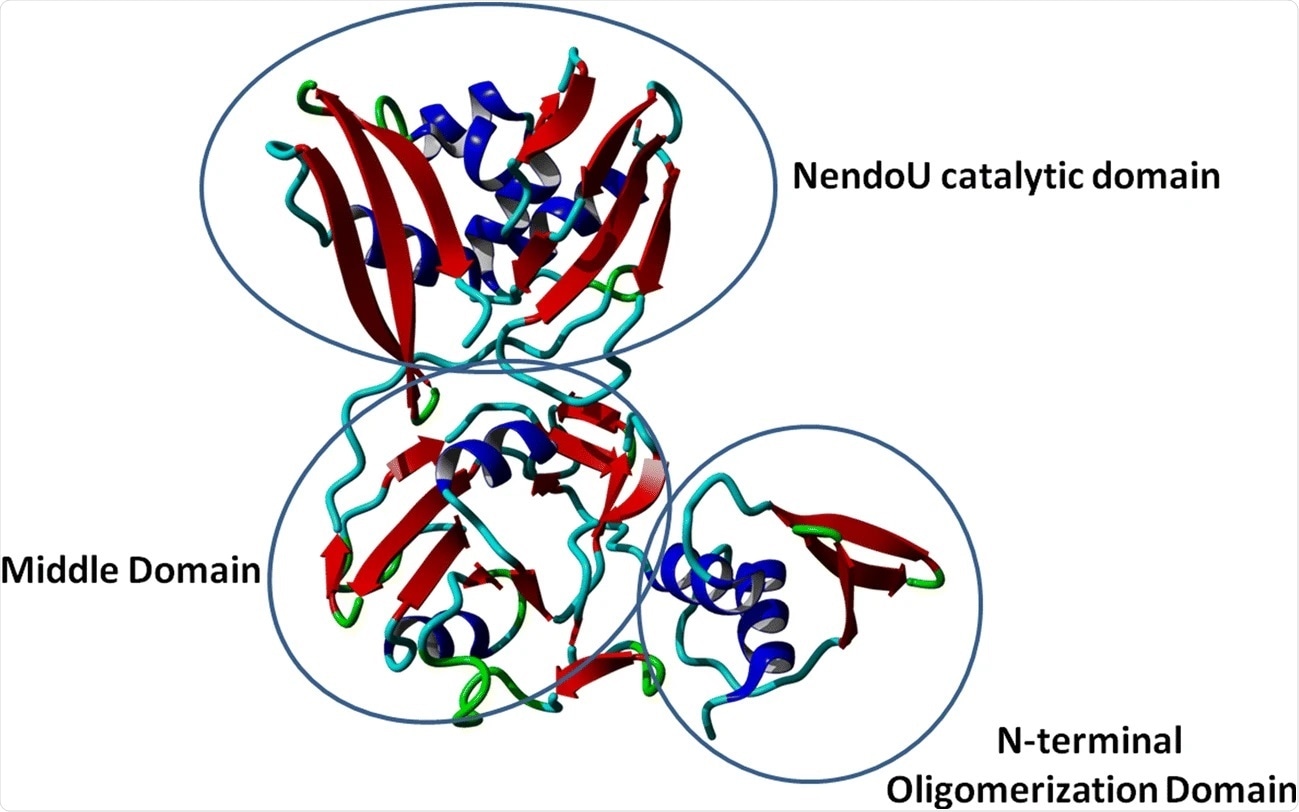
Crystal structure of NSP15 monomer protein. Image Credit: https://link.springer.com/article/10.1007/s00894-021-04750-z/figures/2
NSP15 may be a viral target
The viral ribonucleic acid (RNA) genetic material encodes both its structural proteins, such as the spike and nucleoprotein. However, it also encodes two large polyproteins which are processed to yield 16 different non-structural proteins (NSPs). These are enzymes that are vital for viral replication and further processing of sub-genomic RNA.
However, the function of NSP15 is still unclear. It is also called RNA uridylate-specific endoribonuclease (NendoU), and though it contributes to viral replication, it appears to be not essential for this process. It may also inhibit antiviral cellular responses or may break down viral RNA to evade its detection by the host immune system.
Despite this lack of clarity, NSP15 is required for normal viral function. As a result of this certainty, its crystal structure has been solved in two forms, called the apo and the citrate form. Some aspects of this structure include alpha-helical N-terminal domains, beta-strands and beta-sheets forming the middle domain, and antiparallel beta-sheets forming the C-terminal domain.
The SARS-CoV-2 NSP15 exists as a dimer in the crystal structure but may form hexamers in the active state. The catalytic domain of the NendoU type in the C-terminal domain is part of a widespread and very important RNA processing enzyme.
Earlier studies showed five possible inhibitors of NSP15, namely, dihydroergotamine, glisoxepide, idarubicin, ergotamine, and tasosartan. The current study aimed to design custom compounds to provide broad antiviral activity. This could yield compounds that act like drugs but are already used in people, speeding up the development cycle.
Virtual analysis of natural compounds
Using a mix of natural compounds, from the ZINC library of about 11,000 functional compounds, the researchers virtually screened for high-affinity ligands.
They carried out global docking to the NSP15 macromolecule. This yielded five optimum hits. Of these, the molecules that interacted with NSP15 with the best energy profiles were further studied.
Two molecules, represented as 1776037 and 95372568, showed 21 total interactions, both polar and non-polar. Both had almost identical binding energy, with just 0.6 kcal/mol difference, but the latter has 3 hydrogen bonds compared to only one with the former.
Thus, this was selected as the top hit, and molecular docking simulations were further performed, showing the required degree of accuracy in the protein-ligand docking configurations.
Toxicity prediction
The study also shows that these compounds are the best choices for drug development, as they show the highest efficacy with the lowest side effects. They also fit the Lipinski rule of 5 for a good drug. The Lipinski rule sets guidelines for physicochemical properties of a compound that help decide whether it is likely to be absorbed well when orally administered.
Molecular dynamics (MD) simulations
Further molecular dynamics (MD) studies on five NSP15-ligand complexes showed high equilibration, with all the systems showing sustained stabilization. This was explained by relatively low RMSD (right mean square deviation) energy fluctuations. The best docking score was seen with the compound 1777791, which showed the highest binding affinity in earlier steps.
However, overall, the highest equilibration was seen with compound 1776037.
When hydrogen bonding is examined, the greatest number was seen with NSP15-95372568, but the least with NSP15-25575299 and NSP15-1776037 complexes. Changes in the secondary structure were least in the NSP15-1777791 complex, but in all cases, there is a consistent pattern of change, indicating efficient ligand bonding.
As a result, it is possible that the biological functional effects of 1776037 could be minimized by using it in NSP15 binding along with 95372568, without the risk of adverse effects.
The NSP15 protein does not show any significant changes in the structure after binding with most of these ligands except with compound 1776037. Moreover, the protein is found to be very receptive to the ligand, adapting its conformation to enable better ligand fitting.
However, the NSP15-1776037 complex is the most structurally stable for ligand binding as it has the lowest radius of gyration (Rg). The Rg is a measure of compactness, indicating the overall size of the protein by measuring the distance of the various atoms from the common center of mass.
The other complexes show a slight decrease in the Rg upon ligand binding, indicating that this process helps to stabilize the protein in its stable compact form.
Further free energy analyses show that the NSP15-95372568 complex has the highest interaction with the viral NSP.
What are the conclusions?
Based on the overall results, the study indicates that together with compound 1776037, compound 95372568 offers good prospects for an antiviral drug to effectively counter SARS-CoV-2.
These compounds interact with NSP15 on the active site, especially at residues ARG199, ASN200, TYR279, and ASP297, all on the surface of the protein. These polar and non-polar bonds will be essential to abolishing viral function and thus combating the further transmission of the virus.
The stability and high binding affinity of these compounds have shown them to be promising lead compounds as NSP15 inhibitors, and perhaps both may be used together for more potent inhibition. Further in vitro studies are required to validate these results.
Journal reference:
-
Motwalli, O. et al. (2021). Analysis of natural compounds against the activity of SARS-CoV-2 NSP15 protein towards an effective treatment against COVID-19: a theoretical and computational biology approach. Journal of Molecular Modeling. https://doi.org/10.1007/s00894-021-04750-z. https://link.springer.com/article/10.1007/s00894-021-04750-z