The B.1.1.7 variant of SARS-CoV-2 emerged in the United Kingdom during the ongoing COVID-19 pandemic and was quickly identified as the first major variant of concern that was more transmissible and, arguably, more virulent.
Consequently, due to its infectiousness, it accounted for approximately 90% of COVID-19 cases in Europe and 50% of cases in the US as of May 2020. This variant is characterized by the presence of N501Y mutation in the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein, as well as a handful of other mutations.
Such pervasive presence of this variant opens the door for continuous acquisition of novel mutations, and some of them may prompt the emergence of even more infectious (and potentially more pathogenic) sub-lineages of B.1.1.7.
Naturally, this implies that there is a need for meticulous genomic surveillance in order to be one step ahead of such newly developed mutations in SARS-CoV-2 variants of concern. A research group from the Keck School of Medicine of the University of Southern California in Los Angeles decided to follow the B.1.1.7-M:V70L sub-lineage that has been recently observed with increased frequency.
Chasing mutations with phylogenetic analysis
In short, this study incorporated whole genome sequencing of 2900 samples from Children’s Hospital Los Angeles that were previously found to be positive for SARS-CoV-2 by reverse transcription-polymerase chain reaction (RT-PCR).
Subsequently, phylogenetic analysis has been pursued with complex tools available to researchers (such as the NextStrain phylogenetic pipeline), together with the protein structure prediction of mutant spike glycoproteins.
Furthermore, all candidate mutations were further partitioned by pangolin lineage in order to pinpoint any emerging mutations within the context of a specific lineage (such as B.1.1.7 or B.1.351 variants).
Altering the configuration of SARS-CoV-2
By using the aforementioned methodological approach, this research group has identified a sub-lineage of B.1.1.7 that emerged via sequential acquisitions of M:V70L mutation in November 2020, which was followed by a novel S:D178H mutation initially observed in early February 2021.
As of April 2021, this sublineage comprised 36.8% of all B.1.1.7 isolates found in Washington. More specifically, phylogenetic analysis and transmission inference with Nextstrain implies that its likely origins are either in California or Washington.
Additional structural analysis unveiled that the S:D178H mutation is close to two other signature mutations of B.1.1.7 – Y144del and HV69-70del. Moreover, it is surface exposed and may alter tertiary configuration of N terminal domain (which is a part of spike glycoprotein), affecting the neutralization ability of antibodies.
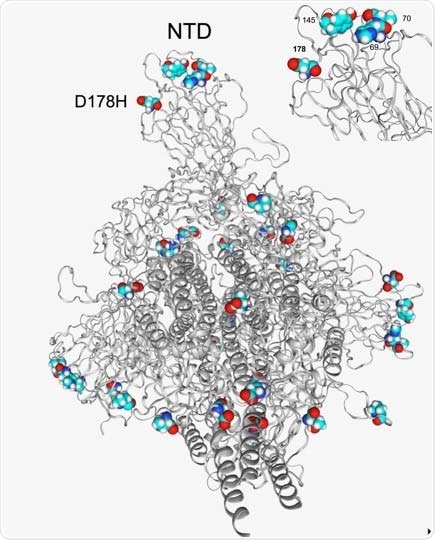
Structural location of the D178H and other spike p 333 protein mutations in the B.1.1.7 lineage. D178H is in close proximity to other amino acids in the NTD that are affected in the B.1.1.7 lineage, HV69-70del and Y144/Y145del (top right).
The potential for ‘vaccine escape’
The observations from this study show that the S:D178H mutation is repeatedly observed but only amplified exponentially in the context of the more pathogenic B.1.1.7 lineage, serving as a valid argument for its fitness.
“These findings highlight the continued importance of active genomic surveillance to monitor the spread of this B.1.1.7-M:V70L-S:178H lineage”, said study authors in this medRxiv paper. “The potential effect of the S:D178H mutation on immunity and ‘vaccine escape’ also warrant further analysis”, they added.
There is a worrying increase in the percentage of B.1.1.7 isolates in the US belonging to this sublineage, as the percentages rose from 0.15% in February to 1.8% in April 2021. To date, this specific sublineage appears to be specific for the US, with reported cases in 31 states (including Hawaii), but continuous surveillance is definitely warranted.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources