Using CRISPR/Cas9 and human intestinal organoids to study genes implicated in the biology of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), researchers from the Netherlands identified a specific serine protease (TMPRSS2) as an attractive pan-coronavirus therapeutic target. Their exciting findings are available in a study currently available on the bioRxiv* preprint server.
The current coronavirus disease (COVID-19) pandemic, caused by the SARS-CoV-2, has exposed earnest weaknesses in our readiness for coronavirus outbreaks and pandemics. More specifically, there are no effective coronavirus antivirals approved for use in humans, while large-scale COVID-19 drug trials came short of proving efficacy – including remdesivir and hydroxychloroquine.
Regarding the latter, we lack an adequate understanding of the basic biology of coronaviruses, as evidenced by the disappointing clinical effect of hydroxychloroquine in humans. Our immediate need should be for the rapid identification of host genes needed for viral replication since only this will allow us to generate treatment interventions more swiftly.
Genetic screens are habitually performed in transformed cell lines that inadequately represent viral target cells in vivo, which results in discoveries that fall short in the translation process to the clinic. Furthermore, substantial differences may occur between individual transformed cell lines.
Conversely, as intestinal organoids are increasingly being used to model human disease (and are also amenable to genetic engineering), a research group led by Dr. Joep Beumer from the Royal Netherlands Academy of Arts and Sciences and University Medical Center in Utrecht, Netherlands decided to use them as non-transformed models to assess genes involved in coronavirus biology.
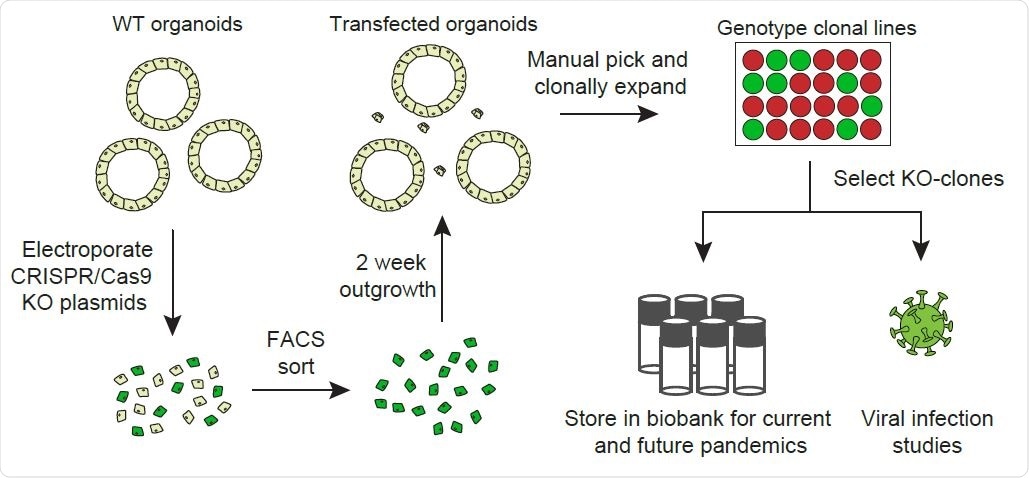
Overview of the workflow of generation of gene knockouts.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A multifaceted use of organoids
In order to discern which host factors can be used as genuine anti-coronavirus therapeutic targets, we have generated mutant clonal intestinal organoids for 19 host genes that have been previously implicated in the biology of coronaviruses. The organoids were also transfected for CRISPR-Cas9 experiments.
“We have chosen to use only intestinal organoids since it is currently not possible to efficiently genetically engineer airway organoids due to limited clonal outgrowth of these cells”, say study authors. “Nevertheless, intestinal organoids express the majority of host factors assessed, including proteases, to a similar level as the airways”, they add.
In addition, they have also performed RNA sequencing of three-dimensional nasal organoids and two-dimensional air-liquid interface differentiated airway organoids. Finally, to show that viral entry occurs through the apical membrane where the main viral receptor is located, the researchers have used mechanically disrupted and intact organoids.
Cell surface protease TMPRSS2 key to replication
In this study, the researchers have confirmed that angiotensin-converting enzyme 2 (ACE2) represents the obligate entry receptor for SARS-CoV-2 and SARS-CoV, whereas a glycoprotein dipeptidyl-peptidase 4 (DPP4) is the entry receptor for MERS-CoV – indicating that accessory receptors may not play crucial roles for these viruses.
Importantly, the replication of SARS-CoV-2 in intestinal organoids did not necessitate the endosomal Cathepsin B/L proteases but characteristically depended on the cell surface protease TMPRSS2. However, other TMPRSS family members were shown to be not essential.
Furthermore, the newly emerging SARS-CoV-2 UK variant B.1.1.7, as well as the original SARS-CoV and MERS-CoV, similarly depended on TMPRSS2, which means this basically represents a pan-coronavirus target. The viral replication was significantly diminished in all TMPRSS2-deficient organoids.
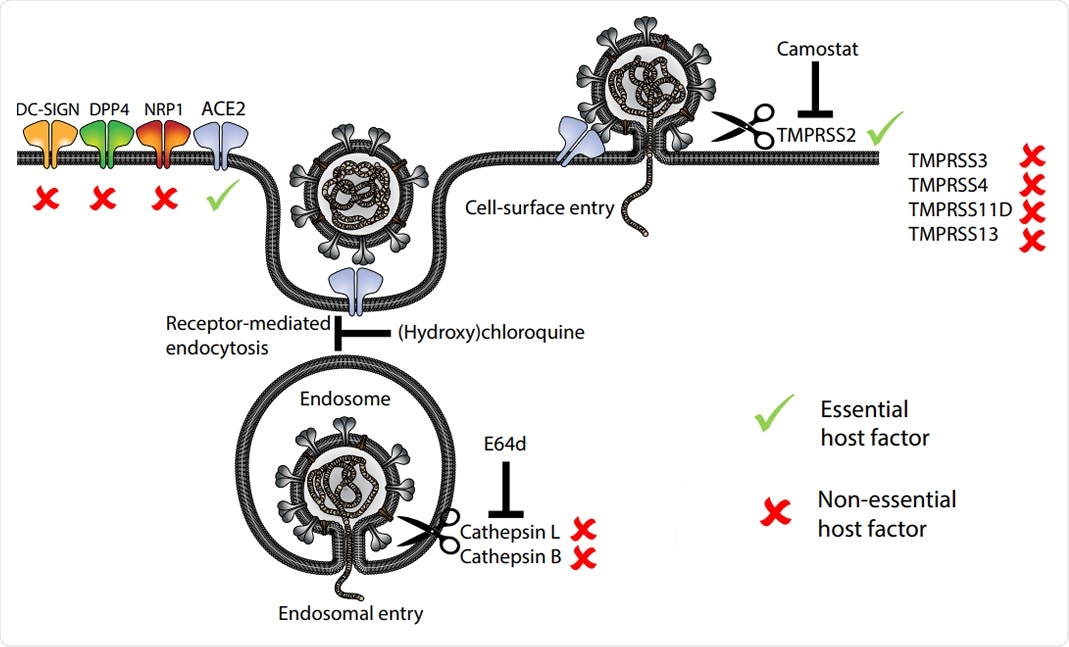
Model of host entry receptors and proteases involved in the entry of SARS-CoV-2. Essential and non essential host factors based on the phenotypes in IO mutants are marked
Towards the identification of novel coronaviral inhibitors
Taking everything into account, these findings imply that manifold TMPRSS genes may be able to mediate cell entry in the state of overexpression; however, at physiological levels in intestinal organoids, it is clear that only TMPRSS2 plays a pivotal role, paving the way for the potential development of highly specific TMPRSS2 inhibitors.
“The high TMPRSS-2 dependency of SARS-CoV found in this study indicates that such inhibitors may well be effective against future SARS-like coronavirus pandemics”, emphasize study authors in this bioRxiv paper. “The observation that TMPRSS2-null mice do not display a visible phenotype implies that such inhibitors may be well-tolerated”, they add.
In conclusion, this study underscores the importance of non-transformed human models for coronavirus research and recognizes TMPRSS2 as a very attractive therapeutic target when compared to a plethora of other genes (e.g. NRP1, NDST1, cathepsin L and/or cathepsin B). Furthermore, future emerging viruses could be identified quickly using this intestinal organoid host factor knockout biobank for rapid discovery of novel therapeutics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Beumer, J. et al. (2021). A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. bioRxiv. https://doi.org/10.1101/2021.05.20.444952, https://www.biorxiv.org/content/10.1101/2021.05.20.444952v1
- Peer reviewed and published scientific report.
Beumer, Joep, Maarten H. Geurts, Mart M. Lamers, Jens Puschhof, Jingshu Zhang, Jelte van der Vaart, Anna Z. Mykytyn, et al. 2021. “A CRISPR/Cas9 Genetically Engineered Organoid Biobank Reveals Essential Host Factors for Coronaviruses.” Nature Communications 12 (1): 5498. https://doi.org/10.1038/s41467-021-25729-7. https://www.nature.com/articles/s41467-021-25729-7.