While the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been curtailed in different regions, the emergence of transmissible variants has led to a peak in the number of cases in April 2021. In the UK, an efficient and widespread genomic sequencing program has made it possible to monitor the rise of such variants.
A new study shows the pattern of the rise of various strains in the UK, as shown by the data collected by the COVID-19 Genomics UK Consortium. This allows the rise and fall of 62 different viral lineages to be traced through over 300 local authorities from September 2020 to April 2021.
The 323 lineages identified during the study period, from the sequenced genomes covering about 5% of all positive tests, were classified into 62 lineages, with a hundred or more genomes in each lineage. The exceptions were variants of concern (VOCs) or variants under investigation (VUIs).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A preprint version of the study is available on the medRxiv* server, while the article undergoes peer review.
Early lineages
In chronological order, the first wide-spreading mutation was D614G, in the spike, which was 20% more transmissible than the original lineage isolated in Wuhan, China. A little later, the B.1.177 strain was isolated in Spain and introduced into multiple European countries.
This was followed by the B.1.351 variant first isolated from South Africa, which contains both the N501Y mutation, which increases its transmissibility, and the E484K mutation that enables it to evade host immunity elicited by earlier strains.
England saw two waves with their peaks in early November 2020 and January 2021. Each triggered a national lockdown.
Up to September 2020, the major circulating lineages were B.1, B.1.1, and B.1.177, making up 100-1,000 genomes each, in different regions in the south and the north of England. These were steadily superseded by B.1.1.7 variants, which showed up to 11% higher growth rates compared to the B.1 or B.1.1 lineages.
B.1.177 lineages also expanded more than B.1 strains until the end of January 2021, in the UK and in Denmark, as it had earlier spread throughout Europe. The reasons are far from clear-cut, as imported infections have not been reported, and the defining A222V spike mutation has no particular growth advantage.
Rise of B.1.1.7
In September 2020, England saw the rise of the B.1.1.7 lineage, first reported in Kent during November and December 2020. This lineage was marked by a 70% increase in transmissibility of this variant relative to B.1.1 and a 45% increase vs. B.1.177.
Thus, the B.1.1.7 sequences increased 20-fold per month compared to B.1.1, and 10-fold higher than B.1.177, accounting for its dominance even when other strains were showing a decline during the period of the second UK-wide lockdown.
This is accounted for by the high transmissibility of the Kent or UK variant, often attributed to the shared N501Y mutation, and it rapidly became the most common variant in over a hundred countries.
B.1.1.7 was observed to drop steeply in prevalence during the third lockdown, over the first few months of the new year, along with most other circulating strains. This lockdown was preceded by increasing regional tiered restrictions as the Kent strain caused ever-increasing cases all over England.
Tier 4 restrictions were associated with a 25% to 50% fall in viral proliferation compared to tier 3. In particular, the mean reproduction number (R) was as low as 0.6 for non-Kent lineages, vs. 0.9 for B.1.1.7. But the R value of the virus continued to be above 1, rising to 1.6 relative to August 2020, with more infectious variants taking over.
This indicates that the pandemic continued to grow. This may have been driven by the Christmas and New Year gatherings.
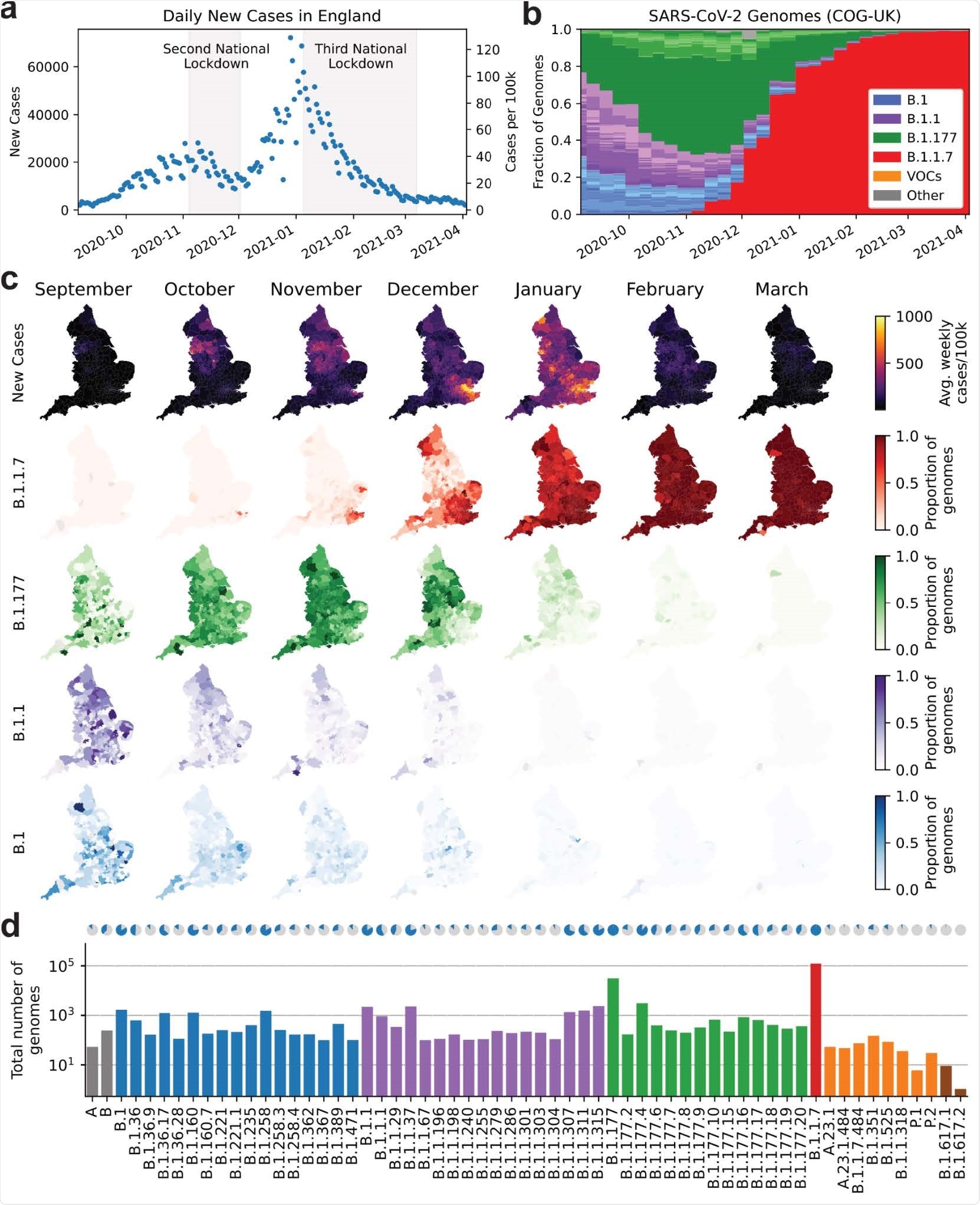
SARS-CoV-2 surveillance sequencing in England between September 2020 and April 2021. a. Positive Pillar 2 SARS-CoV-2 tests in England. b. Relative frequency of 62 different PANGO lineages, representing approximately 5.3% of tests shown in a. c. Positive tests (top row) and frequency of 4 major lineages across 315 English lower tier local authorities. d. Absolute frequency of sequenced genomes mapped to 62 PANGO lineages. Blue areas in the pie charts are proportional to the fraction of LTLAs where a given lineage was observed.
Contraction of cases with the third lockdown
The subsequent lockdown was accompanied by vaccine rollouts, leading to almost 50% immunity by the beginning of March 2021 due to a combination of both natural immunity and increasing vaccination. Daily cases fell from over 72,000 on December 29 to 2,500 at the start of April 2021.
Of the circulating lineages at this time, 99% of cases comprised B.1.1.7, while the B.1.177 strain, which was dominant in November 2020, fell from about 10,000 cases per day in December 2020 to 5 in March 2021. The number of lineages in circulation fell from 137 at the peak to only 22.
The researchers write:
The conclusion that the majority of lineages present in the autumn of 2020 has been eliminated in the spring of 2021 is further supported by the fact that the fraction of positive cases that were sequenced has increased from around 5% in the fall of 2020 to 50% in the week ending April 3, 2021.”
Rise of E484K mutants
Unfortunately, several novel variants containing the spike E484K mutation resisted neutralizing antibodies elicited by the earlier variants and thus began to be noticed at moderately rising levels between December and April.
Whereas VOCs and VUIs made up less than half a percent of infections sequenced for monitoring purposes from January to April 2021, this changed when B.1.617.2 began to rise along with the other E484K lineages. However, these soon peaked and subsided, causing mostly localized outbreaks.
These include the P.1 strain reported first from Brazil, the A.23.1 strain that caused sub-epidemics in Uganda and Rwanda, and B.1.525 from Nigeria, the UK and North America. The B.1.1.318 strain was also prevalent in the UK, as well as the US and Germany.
These may not be more infectious than the B.1.1.7 strain. Most are due to repeated imported infections rather than true community spread, indicating that they are less transmissible even than the growth rates suggest. The exception is an A.23.1 strain that acquired the immune evasion E484K mutation domestically.
Not only did these strains possess only the E484K mutation without other functionally important variants, but they showed no growth advantage – rather, the reverse appears to be true.
Entry of B.1.617 lineages
The last-known introduction is of B.1.617 (in March) and B.1.617.2, which has surged to about 40% of sequenced genomes after its introduction in April 2021. The former has caused a catastrophic second wave in India.
B.1.617 lacks the E484K mutation. B.1.617.1 has an E484Q mutation that is functionally similar to the immune evading E484K spike variant.
B.1.617.2 shows a growth rate that is about 37% higher than B.1.17, indicating that either these are more transmissible or escape immune responses; or that a very high number of cases were introduced within a short period.
Another factor could be the presence of a favorable environment in the localities where these strains increased most rapidly. This means that over time, the growth rate may come down.
And lastly, the increase in B.1.617.2 (which now makes up 40% of sequences) is counteracted by the fall in B.1.1.7 cases. Yet B.1.617.1 did not show such a rapid rise, with 3-4 sequenced genomes for each introduction vs. 10 with the former strain.
What is the implication?
This data offers “a stark reminder that viral evolution can take unexpected paths and that rapid genomic surveillance remains as important as ever.” Moreover, it reminds us that even interventions that worked once may not be so effective with newer and more infectious variants.
It is unlikely that global eradication is possible at least in the short to mid term. Therefore a global perspective on controlling and surveilling the virus is essential,” conclude the researchers.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Vohringer, H. S. et al. (2021). Genomic reconstruction of the SARS-CoV-2 epidemic across England from September 2020 to May 2021. medRxiv preprint. doi: https://doi.org/10.1101/2021.05.22.21257633, https://www.medrxiv.org/content/10.1101/2021.05.22.21257633v2
- Peer reviewed and published scientific report.
Vöhringer, Harald S., Theo Sanderson, Matthew Sinnott, Nicola De Maio, Thuy Nguyen, Richard Goater, Frank Schwach, et al. 2021. “Genomic Reconstruction of the SARS-CoV-2 Epidemic in England.” Nature, October. https://doi.org/10.1038/s41586-021-04069-y. https://www.nature.com/articles/s41586-021-04069-y.