Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019 (COVID-19) pandemic, is highly transmutable and infectious. In order to detect different stages of the viral infection appropriately, various diagnostic methods are required.
A group of international researchers has investigated multiplexed detection of COVID-19 with single-molecule technology. Using a single-molecule enzyme-free assay for multiplexed detection has several benefits, including flexibility and the ability to quantify low-volume samples effectively. Researchers developed a platform for detecting the virus directly from a patient's sample as well as the immune response through antibodies such as IgG and IgM.
A preprint version of the research paper is available on the medRxiv* server while the article undergoes peer review.
Current Diagnostic Tools
The current diagnostic tests used for viral infections include real-time reverse transcription-polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA). These tests require multiple steps and involve enzymatic-based signal amplification; the length of time for these diagnostic tests can be quite long, and this can make using these a little inconvenient to depend upon during a pandemic.
The pandemic has required dependence on healthcare systems like never before. To ensure that individuals are safe to return to work or engage in other social activities after presenting with symptoms, COVID-19 PCR tests are required in high quantities within a short period of time. Due to this, new diagnostic methods may be more effective during a pandemic and in handling a viral infection such as SARS-CoV-2.
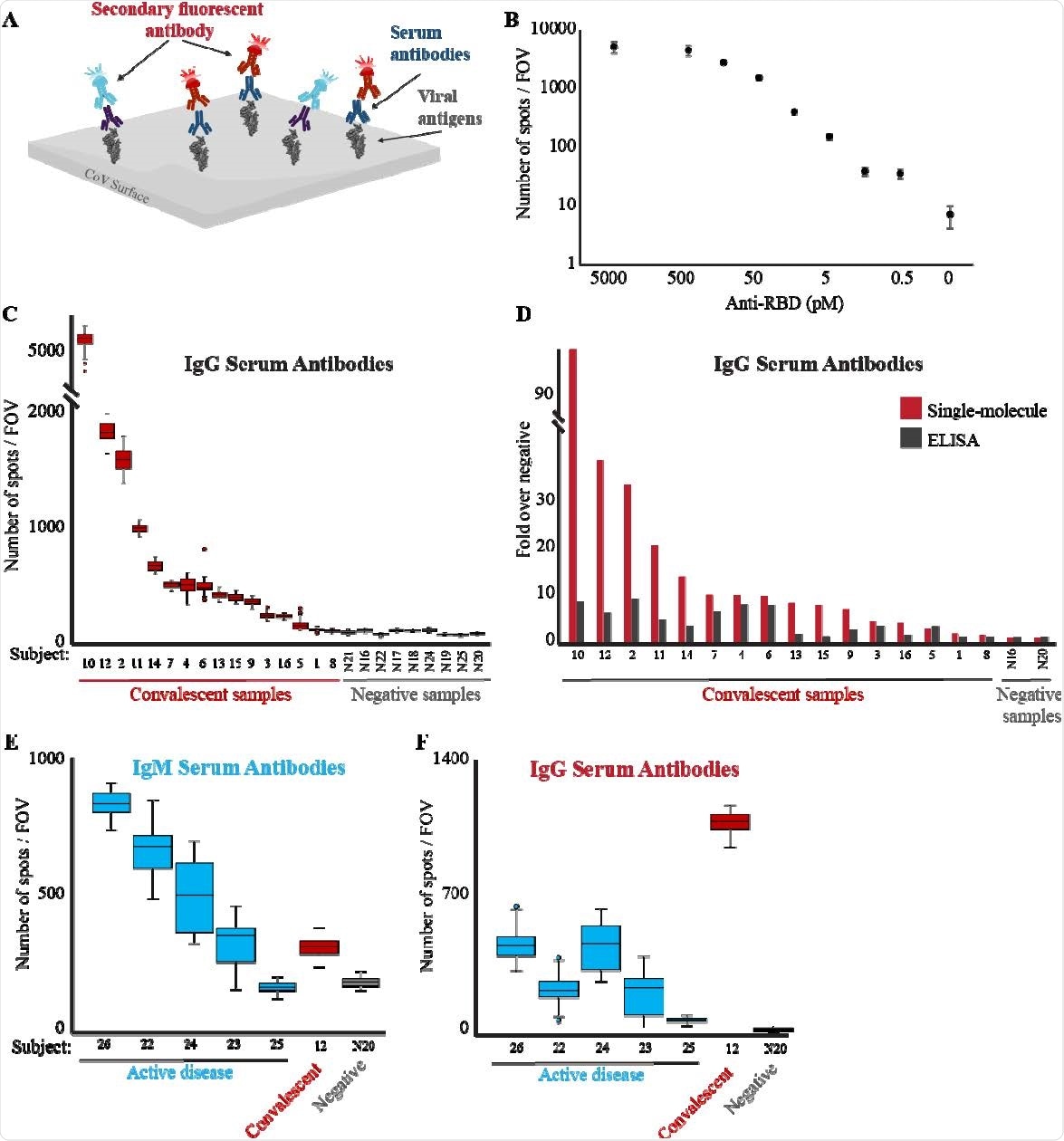
Single-molecule detection of anti-RBD antibodies. (A) Scheme of the serological diagnostic test. Serum samples are incubated with biotin-conjugated viral antigen (RBD) and loaded on a PEG-coated, streptavidin activated coverslip. Multiplex of fluorescently-labeled anti-human IgG (red) and IgM (light blue) antibodies are added to the flow cell and imaged. (B) Human anti-RBD antibodies at the indicated concentrations were incubated with biotin-RBD, and detected by fluorescently-labeled anti-human IgG antibodies. The Antibodies LoD is at picomolar concentrations. Both axes are in logarithmic scale, and the no anti-RBD antibody data point is not to scale. (C) Serum samples from either convalescent or not-infected subjects were diluted 1:2500 and analyzed as described in B to detect the presence of anti-RBD IgG antibodies in the subjects’ serum. The box plot shows the number of spots per FOV for all the FOV imaged for each sample. Median values of each group were compared by t-test, p-value < 0.05. (D) Comparison between single-molecule and ELISA detection of anti-RBD antibodies. Single-molecule imaging and ELISA against anti-RBD antibodies were conducted on the same samples. Signals from each assay were normalized compared to the negative serum samples. Single-molecule imaging provides greater sensitivity and dynamic range in detecting anti-RBD antibodies in serum. (E-F) Serum from subjects with an active COVID-19 disease (blue), convalescent (red), or not-infected (gray) subjects, were diluted 1:2500, incubated with biotin-RBD and loaded on a streptavidincoated surface. Fluorescently labeled anti-human IgM (E) or IgG (F) antibodies were imaged and quantified.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
New Diagnostic Approaches
The development of novel diagnostic methods has risen in response to the time-consuming, multistep, and costly demands within current diagnostic methods. New diagnostic methods that have been developed, include: reverse transcription coupled with nanopore sensing, isothermal amplification, CRISPR-based methods, next-generation sequencing-based approaches, as well as research into improving RT-PCR timing through plasmonic thermocycling.
Although these new diagnostic methods aim to improve time for tests, as well as cost and accessibility, they still mostly rely on enzymatic processes.
Single-based Imaging Technology
Single-based imaging technology has improved over the years from having higher throughput sequencing technology to being sensitive enough to detect proteins.
Along with other researchers, the authors of this paper have shown that Total Internal Reflection Fluorescence (TIRF) microscopy is able to detect single fluorophores attached to a solid surface while providing spatial and spectral multiplexing and quantitative detection of molecules.
Within this research paper, the scientists present proof-of-concept for streptavidin-biotin surface capturing, with fluorescent labeling, in order to detect viral RNA and anti-viral serum antibodies through single-molecule imaging. Tests on samples showed that these diagnostic methods are likely to be more efficient than conventional enzymatic methods for detecting viral infections due to their high scalability and minimal dependency on enzymes.
The researchers developed the detection of RNA by TIRF microscopy through the following three steps. The first step includes in-tube hybridization, which consists of viral RNA being hybridized with two types of DNA probes, including probes labeled with biotin and detection probes labeled with a fluorophore.
The second step is immobilization, where samples are added to a flow cell with a streptavidin-coated coverslip, which enables the hybridization complexes to be captured by the biotin-streptavidin interaction.
The third and final step includes imaging, where the complexes are able to be imaged by the TIRF microscope, and this method enables each spot imaged to correspond to a single molecule of viral RNA.
Significance for COVID-19
Using single-based imaging technology, the researchers developed a method to detect viral RNA that could be used to detect the SARS-CoV-2 virus at various stages of its life cycle within the body. The researchers were able to develop a three-step approach utilizing TIRF microscopy in order to image and detect single molecules of viral RNA.
Although the novel diagnostic approach detected antibodies more effectively than the classical ELISA assay, the sensitivity of single-molecule hybridizations may not be as effective as amplification-based PCR tests. However, by developing the novel method with a higher level of sensitivity through single-molecule kinetic fingerprinting, this limitation can be overcome.
Besides providing a database of probes that can be used to detect SARS-CoV-2, the design is flexible enough to detect additional pathogens.
Due to the variety of variants that emerged during the recent COVID-19 pandemic, this single-molecule genetic test offers the greatest potential for multiplexed detection of more than one variant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Furth, N., Shilo, S., Cohen, N., Erez, N., Fedyuk, V., Schrager, A., Weinberger, A., Dror, A., Zigron, A., Shehadeh, M., Sela, E., Srouji, S., Amit, S., Levy, I., Segal, E., Dahan, R., Jones, D., Douek, D. and Shema, E., 2021. Multiplexed Detection of COVID-19 with Single-Molecule Technology, https://www.medrxiv.org/content/10.1101/2021.05.25.21257501v1
- Peer reviewed and published scientific report.
Furth, Noa, Shay Shilo, Niv Cohen, Nir Erez, Vadim Fedyuk, Alexander M. Schrager, Adina Weinberger, et al. 2021. “Unified Platform for Genetic and Serological Detection of COVID-19 with Single-Molecule Technology.” Edited by Ruslan Kalendar. PLOS ONE 16 (7): e0255096. https://doi.org/10.1371/journal.pone.0255096. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0255096.