The emergence of numerous severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern with enhanced transmissibility are increasingly being reported around the world, though these strains often result in fewer hospital admissions and less severity of illness and appear to impact vaccine efficacy minimally.
Mutations to the spike protein and receptor-binding domain are responsible for these changes in immunogenic profile, and in a paper recently uploaded to the open access journal Antibiotics by Ambrose et al. (May 6th, 2021) these mutations are correlated with various disease parameters by in silico methods, finding that particular features are responsible for the increased infectiousness and lesser disease severity observed in these lineages.
How was the study performed?
The group obtained SARS-CoV-2 genomic data from international repositories and utilized software to translate the genome into protein sequences, which were then run against B-cell and T-cell epitope prediction tools to identify peptide sequences that would bind with the translated spike proteins with high affinity, promoting removal by these immune cells.
Four SARS-CoV-2 variants of concern were selected in particular for detailed analysis, each known to bear a number of mutations to the spike protein that promote transmissibility.
The D614G mutation has been identified in several SARS-CoV-2 lineages, firstly in February 2020, with the N501Y mutation being identified in December 2020, which when both present were distinctive features of the Alpha variant B.1.1.7. The N501Y mutation was later found to have independently evolved in the Beta B.1.351 around the same time. Gamma P.1, and Delta B.1.617.2 variants also bear numerous mutations to the spike protein, many in common with other lineages, though it is as yet unclear whether these mutations have independently or subsequently evolved.
A phylogenetic tree was constructed computationally for the five strains of SARS-CoV-2, SARS-CoV-1, bat coronavirus, and MERS, demonstrating the close genetic relationship between the multiple variants of SARS-CoV-2 and their direct bat coronavirus ancestor. SARS-CoV-1 is more closely related to both than MERS, though it is evident that MERS also shares a common ancestor with each.
Identifying high-affinity epitopes
T-cell epitopes were identified in silico on the spike protein sequences of each variants, each generating differing numbers of complementary peptides. Within the later variants the highest-affinity peptides exhibited greater antigenicity than those generated against the wildtype strain. However, all epitopes were found to be 100% conserved from wildtype through to each later lineage.
Interestingly, antibodies generated against earlier lineages of SARS-COV-2 have been demonstrated by numerous studies to be less effective in neutralizing strains that have emerged more recently, while immunity developed against the newer lineages is often applicable to earlier variants. This may, at least in part, be due to the highly conserved high-affinity T-cell epitope sites identified in this study and also explain why vaccines remain effective against the newly emerging variants.
Potential linear B-cell epitopes were also identified on the spike protein computationally. The B.1.1.7 and B.1.351 variants were predicted to bond with a more significant number of antigenic peptides. Again, the highest affinity peptides differed amongst the variants, though they were found to be less conserved than in the case of T-cell epitopes. The P.1, and B.1.617.2 variants again bore the most similarity in the top-scoring peptides, though interestingly the B.1.351 and B.1.617.1 variants were more similar in this case, and distinct from the B.1.1.7 and B.1.429, B.1.427 variants.
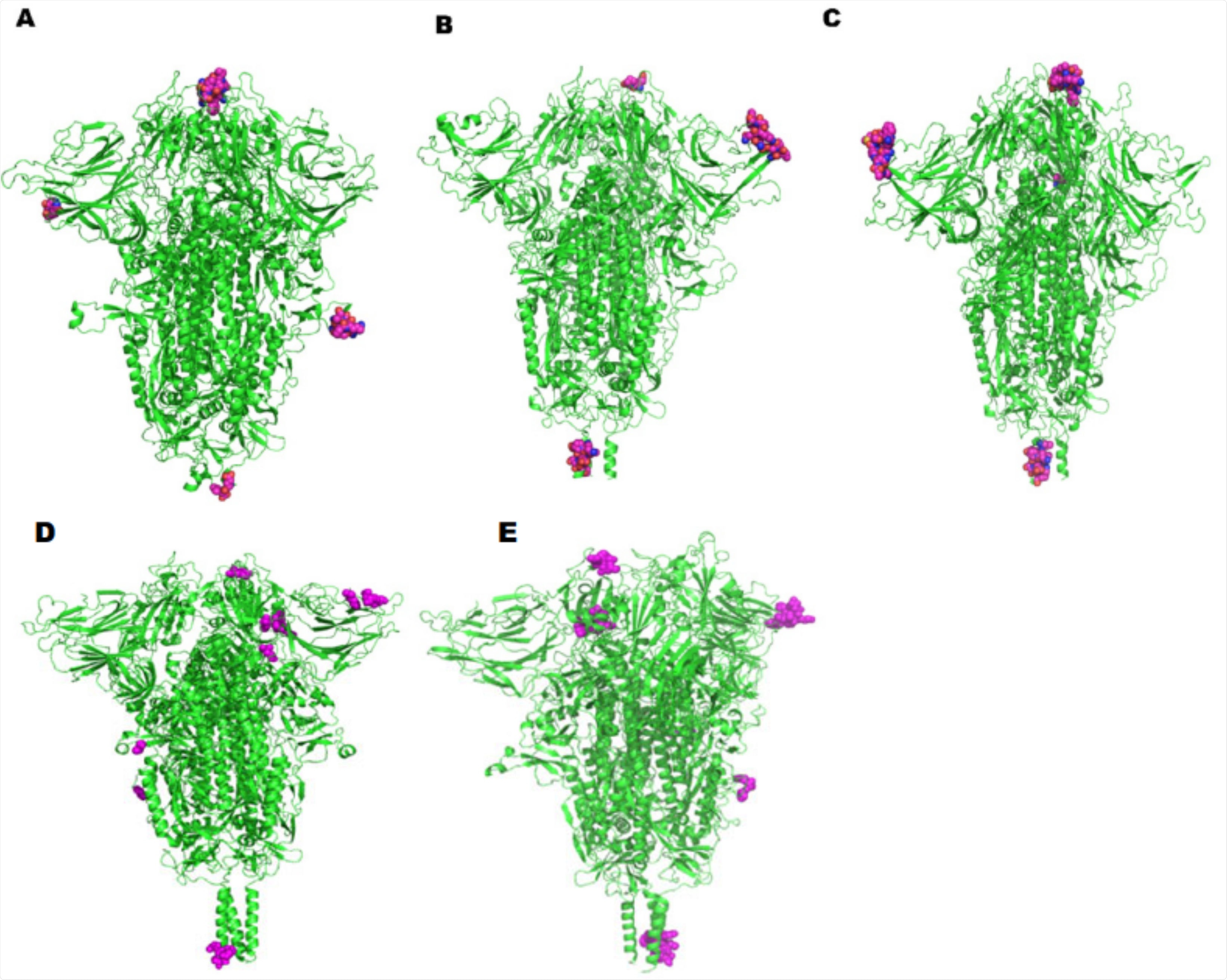
Representations of potential discontinuous B-cell epitopic regions mapped onto the spike protein of the SAR-SCoV-2 variants: (A) Wuhan original L strain, (B) England - B.1.1.7 Alpha, (C)USA - B.1.429, B.1.427 Epsilon, (D) India - B.1.617.1 Kappa, and (E) South Africa - B.1.351 Beta, highlighted as spheres.
Conformationally appropriate spike proteins were then examined from each lineage instead of linear chains, and the highest affinity peptides towards each lineage spike protein were assessed.
The original L strain bears a distinctive GKQ motif at positions 181-183, while the B.1.1.7 and B.1.429, B.1.427 variants both bear the sequence GTNG at positions 72-75. The B.1.617.1 variant alone exhibits an NLK sequence at positions 460-462, with both this and the B.1.351 variant bearing an NHTS sequence at positions 1158-1161. The group note that all of the highest-ranked B-cell and T-cell epitopes against the spike proteins of each later lineage exhibit greater antigenicity than any towards the wildtype original L strain, possibly indicating the reason for lower disease severity observed in these later variants.
The binding conformations of the spike protein of each lineage while bound with antibodies were also examined in silico, finding all to be stable past 6,000 picoseconds. The protein-peptide complex of the B.1.1.7 and B.1.617.1 variants showed the least conformational fluctuation, while the B.1.429, B.1.427 variants fluctuated somewhat and then became stable after 6 nanoseconds, while the B.1.351 variant moderately deviated for 8,000-1,000 nanoseconds before stabilization. The wildtype original L strain showed both more conformational deviation and longer time to stabilization than any of the later lineages, suggesting that mutations in the binding region have brought about conformational change that increases binding affinity while better maintaining rigidity in the complex, subsequently improving the ability of neutralizing antibodies to target the later lineages, and potentially attenuating the severity of disease. As antibodies take several days or weeks to reach sufficient levels following infection, transmissibility is unaffected.