The study findings reveal that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection results in abnormal modification of some lung proteins, which are subsequently transported to the brain via exosomes. In the brain, these modified lung proteins disrupt the normal functioning of proteins that are related to Parkinson’s disease.
Background
COVID-19 caused by SARS-CoV-2 is associated with a wide variety of clinical presentations, including pulmonary, cardiovascular, renal, hepatic, gastrointestinal, and brain pathologies. Several of these symptoms, especially neurological symptoms, have been found to significantly associate with post-COVID complications. In this context, studies have suggested that the long-term presence of neurological symptoms in COVID-19 patients can eventually lead to the development of neurodegenerative diseases and dementia.
Recently, three studies have separately claimed the development of Parkinson’s disease (PD) in 3 COVID-19 patients who had no family history of PD.
Although the exact etiological connection between COVID-19 and post-COVID PD has not been established, it has been hypothesized that direct damage to the nigrostriatal system due to vascular insults, neuroinflammation-mediated death of nigral dopamine neurons, or direct SARS-CoV-2-mediated damage of the brain could be the potential reasons behind the development of PD in COVID-19 patients. However, these hypotheses need further scientific validation.
In the current study, the scientists have evaluated the molecular mechanism responsible for the development of PD in individuals infected with SARS-CoV-2.
Study hypothesis
As a probable mechanism, the scientists hypothesized that damage that occurs in the SARS-CoV-2-infected lungs is propagated to the central nervous system to induce the pathogenesis of PD. They established a simplistic mechanism in support of their hypothesis.
The mechanism describes that after entering human cells, SARS-CoV-2 releases its RNA and proteins into the lungs, which subsequently interact with human proteins. Through this interaction, viral proteins trigger post-translational modification of human proteins, leading to their abnormal expression, misfolding and incorrect assembly, altered localization, and aberrant functioning.
The modified human proteins are then transported from the lungs to the central nervous system via exosomes. They interact with and alter the functioning of proteins related to PD.
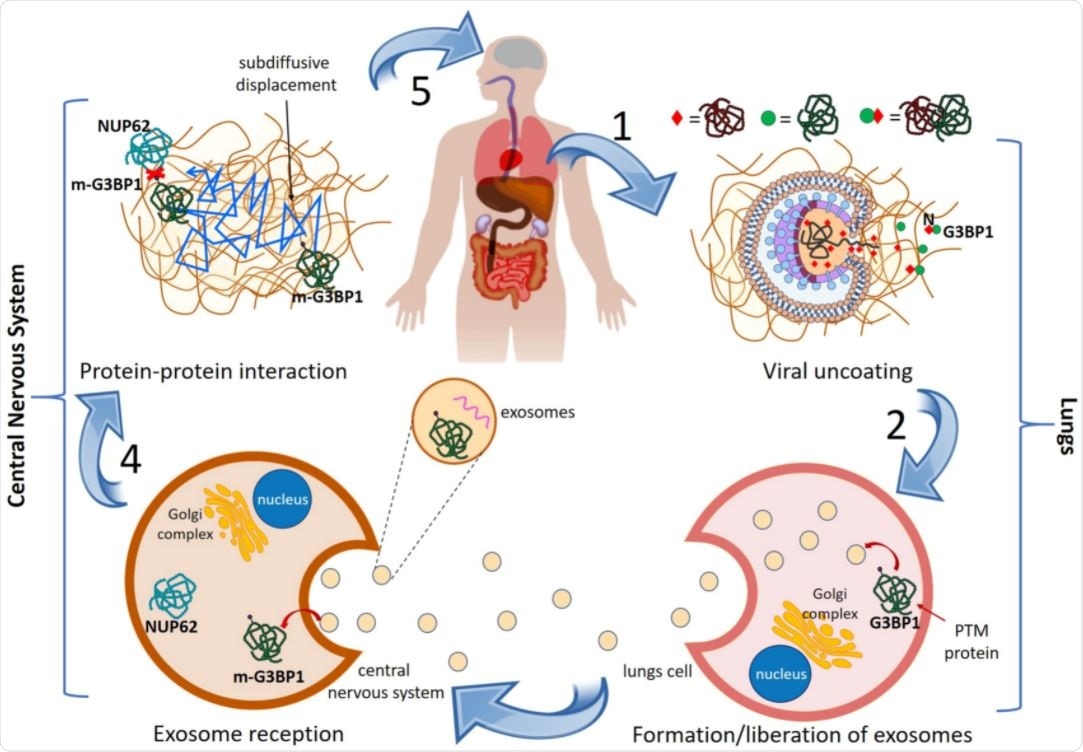
General mechanism of lungs-to-CNS cascade giving rise to PD in COVID-19 patients. See text for explanation.
Identification of SARS-CoV-2-modified human proteins
To identify potential host proteins, the scientists initially started with 332 human proteins that are known to directly interact with SARS-CoV-2 proteins.
With further screening, they specifically selected a set of 278 SARS-CoV-2-interacting human proteins that are highly expressed in the lungs and eventually encapsulated in the exosomes.
In a separate screening, they identified 44 proteins that are vulnerable to PD. By analyzing the interactions between SARS-CoV-2-interacting proteins and PD-vulnerable proteins, they finally identified a set of 24 proteins that directly undergo protein-protein interaction with at least one PD-vulnerable protein.
In other words, these newly identified 24 proteins, termed as “perturbators” by the scientists, possess specific characteristics, including direct interaction with a SARS-CoV-2 protein; significantly high pulmonary expression; exosomal encapsulation; and direct protein-protein interaction with PD-associated/vulnerable proteins.
The interactome between COVID-19 and Parkinson’s disease
The extensive bioinformatic screening performed in the study led to the identification of 44 PD-vulnerable proteins, which are potentially modified by 24 identified perturbators. The scientists identified two proteins, PRKACA and PRKAR2B, as both perturbators and PD-vulnerable proteins. These proteins are highly expressed in the lungs, as well as in the brain.
Furthermore, they formed a protein-protein interaction network of all identified PD-vulnerable proteins and their perturbators. Among various PD-vulnerable proteins, Rab7A and NUP62 showed the highest number of potentially perturbated interactions. Rab7A plays a vital role in autophagic degradation and removal of alpha-synuclein protein aggregates from brain cells. Excessive accumulation of alpha-synuclein aggregates is one of the significant hallmarks of PD.
Regarding NUP62, the scientists explained that like other viruses, SARS-CoV-2 inhibits autophagy in human cells by forming ubiquitinated protein aggregates. This, together with excessive exosome formation, can result in the accumulation of NUP62 and alpha-synuclein protein aggregates.
Four proteins including NUP88, NUTF2, RAE1, and G3BP1 were identified as potential perturbators of NUP62 in the study. These perturbators are known to interact with SARS-CoV-2 proteins Nsp9, Nsp15, Orf6, and nucleocapsid, respectively. Moreover, abnormal functioning of NUP62 is known to associate with the development of PD.
Alternatively, studies have shown that sequestration of G3BP1 and G3BP2 by the viral nucleocapsid protein triggers NUP62-induced aggregation of alpha-synuclein, leading to the development of PD.
Study significance
The study identifies 24 highly expressed proteins in the human lung and can be modified by SARS-CoV-2 proteins. These modified proteins (perturbators) are transported from the lungs to the brain via exosomes to disrupt the normal physiological functioning of 44 PD-vulnerable proteins.
The entire phenomena may collectively explain the molecular mechanism responsible for the development of PD in COVID-19 patients.