Recent studies suggest that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), like other coronaviruses, hijacks various host cell machinery such as autophagy for replication and pathogenesis. Double membrane vesicles formed during the initiation of the autophagy cascade serve as a platform for the assembly of viral replication complexes and enable RNA synthesis. The use of autophagy inhibitors such as chloroquine and hydroxychloroquine were initially used as a potential strategy for the treatment of COVID-19 patients. However, their use was later questioned due to high cytotoxic effects.
There is an urgent need for drugs that can block initial viral infection and spread to control the COVID-19 pandemic in the absence of a fully effective treatment or vaccine. While research has shown a strong connection between autophagy machinery and viral pathogenesis, it is unclear if targeting the autophagy pathway will help fight against SARS-CoV-2 infection.
Researchers recently reviewed current knowledge on links between autophagy and coronaviruses and how this knowledge can be used to repurpose autophagy modulators as potential treatment options for COVID-19. The review is published in the journal Frontiers in Microbiology.
Autophagy and viral infections
Autophagy is a cellular catabolic mechanism that determines the degradation of undesirable protein molecules and recycles the degraded materials. Autophagy was first described in yeasts and is evolutionarily conserved in many higher life forms. It helps in maintaining cell homeostasis and resists invading cellular pathogens.
Several studies on the interplay between viruses and autophagy have shown that many viruses have evolved over time to escape autophagy-mediated degradation and sometimes even use autophagy mechanisms to assist their own replication. As a consequence, many autophagy-modulating agents have antiviral properties and are currently being tested for therapeutic interventions.
Although the link between coronavirus infection and autophagy remains unclear, current knowledge shows that coronaviruses interact with the many components of autophagy machinery to facilitate viral replication. One obvious clue showing this connection is the induction of characteristic double-membrane vesicles (DMVs) or autophagosomes and the reliance of coronaviruses on DMVs to form RNA-replication complexes and infectious viral particles.
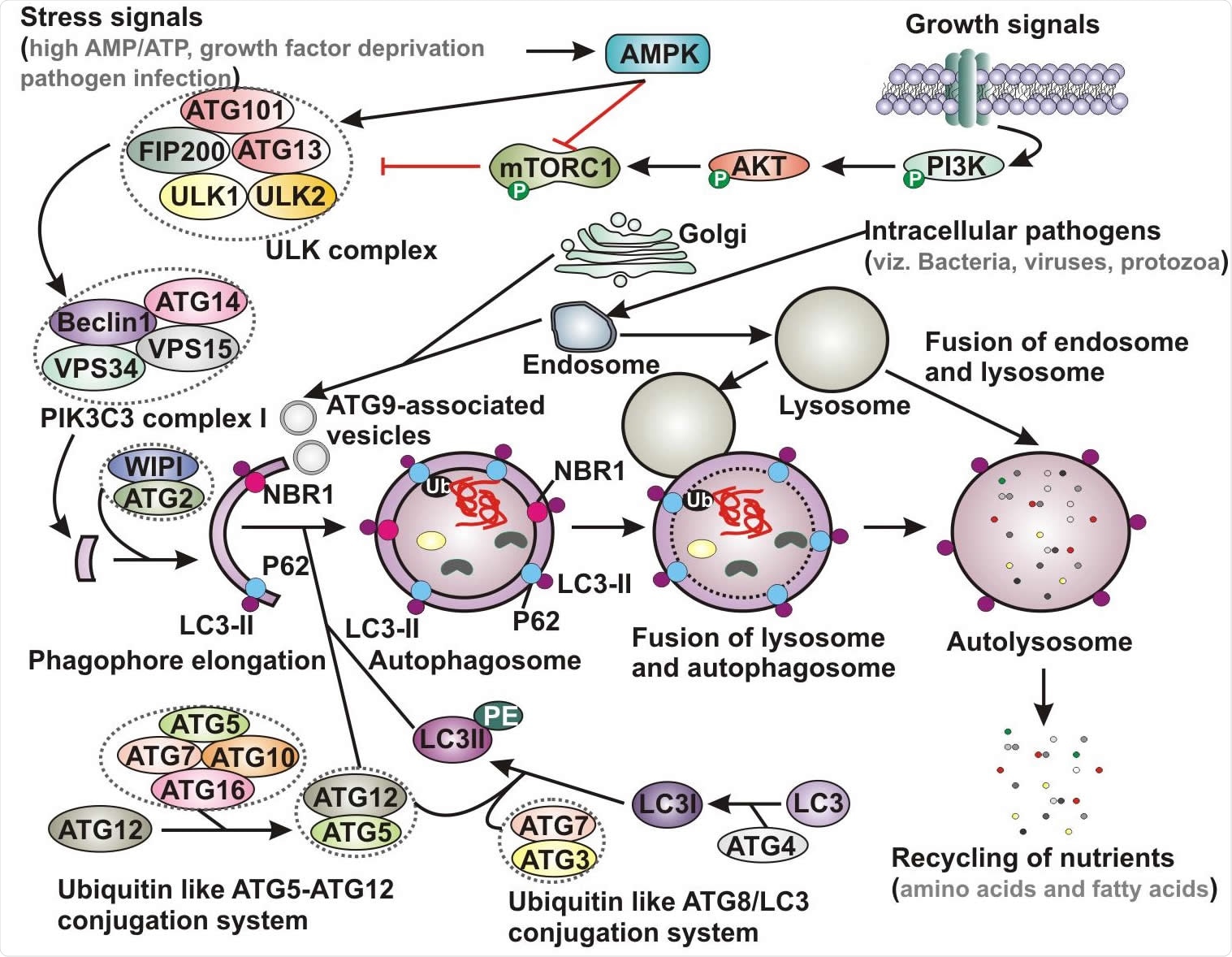
Schematic representation of basic autophagy pathway. Upon deprivation of growth factors, increase in AMP level and pathogen infection lead to AMPK activation and subsequent inhibition of mTORC1 function. In contrast, in the presence of growth signals PI3K-AKT signaling pathway activates mTOR. Inhibition of mTORC1 results in activation of ULK complex, which phosphorylates Beclin-1, leading to VPS34 activation and initiation of phagophore formation. ULK functions in a complex with ULK1, ULK2, FIP200, ATG13 and ATG101, while VPS34 function within the PIK3C3 complex containing its regulatory subunit, VPS15, ATG14 and Beclin-1, which further recruits to WIPI and ATG2 for phagophore elongation. Several ATG proteins engage two evolutionarily conserved ubiquitin-like conjugation systems - ATG12-ATG5 and phosphatidylethanolamine (PE)-conjugated LC3 (LC3-II) targeted to the pre-autophagosomal membrane. In the ATG12-ATG5 conjugation system, the complex further interacts with ATG16, where ATG7 functions as an E1-like enzyme and ATG10 factions as an E2-like enzyme. In the other system, LC3 is first cleaved by a cysteine protease ATG4 to generate LC3-I, which is further conjugated with PE to form membrane bound LC3-II facilitated by ATG7 and ATG3. The cytoplasmic damaged cargo is then ubiquitinated, captured by adaptor molecules - p62 or NBR1 and subsequently delivered to the phagophore membrane. Matured autophagosome then fuses with endolysosomal vesicles forming an autolysosome, where the cargo is degraded and provide nutrients. AMPK, AMP activated protein kinase; mTORC1, mammalian target of rapamycin complex 1; PI3K, phosphatidylinositol 3-kinase; PIK3C3, Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3; LC3, microtubule-associated protein 1 light chain 3; Ub, ubiquitin; NBR1, neighbor of BRCA1 gene 1.
Compounds affecting autophagy pathway may serve as alternative treatment approaches to SARS-CoV-2 infection
The researchers discussed some compounds that directly or indirectly affect the autophagy pathway and may be used as an alternative treatment approach to combat SARS-CoV-2 infection. The compounds include rapamycin / sirolimus, a serine-threonine kinase that serves as a central regulator of cell metabolism, survival, and immune regulation. It is an FDA-approved immunosuppressive drug that inhibits the mammalian target of rapamycin complex 1 (mTORC1) activity. Evidence from studies shows that mTORC1 plays an important role in regulating viral replication.
Everolimus is another compound having antiviral effectiveness and is an FDA-approved second-generation sirolimus derivative. Trehalose, a natural, non-reducing disaccharide found in plants and microorganisms, shows cytoprotective properties against many metabolic disorders and neurodegenerative diseases by activating autophagy.
Another compound, valinomycin, is a cyclodepsipeptide antibiotic reported as a compelling anti-coronavirus compound based on screening of over 10,000 small molecule drug candidates that block SARS-CoV replication.
Several other novel small molecule compounds are currently being tested and more randomized controlled trials (RCT) are urgently needed to confirm that these drugs do not have any severe toxicity issues. A total of 3,807 clinical trials on COVID-19 are in progress so far, and most of the compounds studied are repurposed drugs.
Other research also shows that drugs specifically inducing autophagic flux may have potent antiviral activities in patients with COVID-19. According to the authors, further in-depth studies are warranted to accurately describe the role of autophagy in coronavirus replication and allow the development of new treatment strategies that modulate the autophagy cascade.
Study highlights the importance of pharmacogenomics studies in boosting clinical benefits
Several treatment approaches focusing on modulating autophagy-endocytic pathway have been studied, and many are currently undergoing trials to control the ongoing COVID-19 pandemic. This work highlights the importance of pharmacogenomics studies in boosting clinical benefits and success rates of drug repurposing in patients with COVID-19.
Locating genetic determinants of viral pathogenesis may offer a unique opportunity for drug repurposing in personalized COVID-19 treatment. The authors strongly believe that autophagy inducers including sirolimus, everolimus, valinomycin, and trehalose are promising alternative COVID-19 treatment strategies.