The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) across the United States during 2020 has been said to have occurred in three “waves” or “phases,” characterized by spikes in the number of reported new cases and a roving geographical distribution.
A number of SARS-CoV-2 lineages with higher transmissibility compared to wildtype were identified during this time, known as variants of concern, raising concerns about the rate of virus mutation and what this means for acquired and engineered immunity.
In a research paper recently uploaded to the preprint server medRxiv* by Capoferri et al. (June 4th, 2021), the genetic diversity of SARS-CoV-2 through each phase is examined in detail using publicly available genomic data available from before 2021, highlighting the need to continuously track and assess the evolution of the virus to ensure the future efficacy of the currently available vaccines.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The phases of COVID-19 spread
SARS-CoV-2 was introduced to the US from Europe and Asia in winter 2019, with cases rising rapidly until spring 2020, referred to as phase 1.
The northeast was affected particularly, with many community transmissions occurring in this short space of time. Phase 2 began in summer 2020, this time with the south-western USA bearing a greater burden of cases as non-pharmaceutical interventions were beginning to be relaxed.
The mid-west saw the earliest surge in cases at the beginning of phase 3 in fall 2020, though cases were on the rise nationwide before widespread vaccine distribution began in early 2021.
The authors note some discrepancies in the distribution of cases across the USA and the available SARS-CoV-2 genomic sequences. For example, while the south bore the majority of cases overall, most genomic sequences were obtained from the west.
In total, only 1.2% of all reported cases in the country throughout 2020 had a corresponding viral sequence, compared with 8.1% in the UK and 6.2% in Australia. The median time from sample collection to full genomic sequence acquisition is around 100 days. Thus many samples from the latter stages of phase 3 were unavailable to the group at the time of writing, though they note that the overall sequencing rate has improved in 2021 from the 2020 levels.
Tracking SARS-CoV-2 clades
GISAID is an international organization that monitors influenza and now SARS-CoV-2, providing open access genomic data on the viruses. They categorize SARS-CoV-2 clades and lineages based on differences in genetic sequence, assigning them letter symbols for easy identification. The earliest GISAID-assigned clades were: G, GH, GR, S, L, and V, each of which was identified in the USA during phase 1.
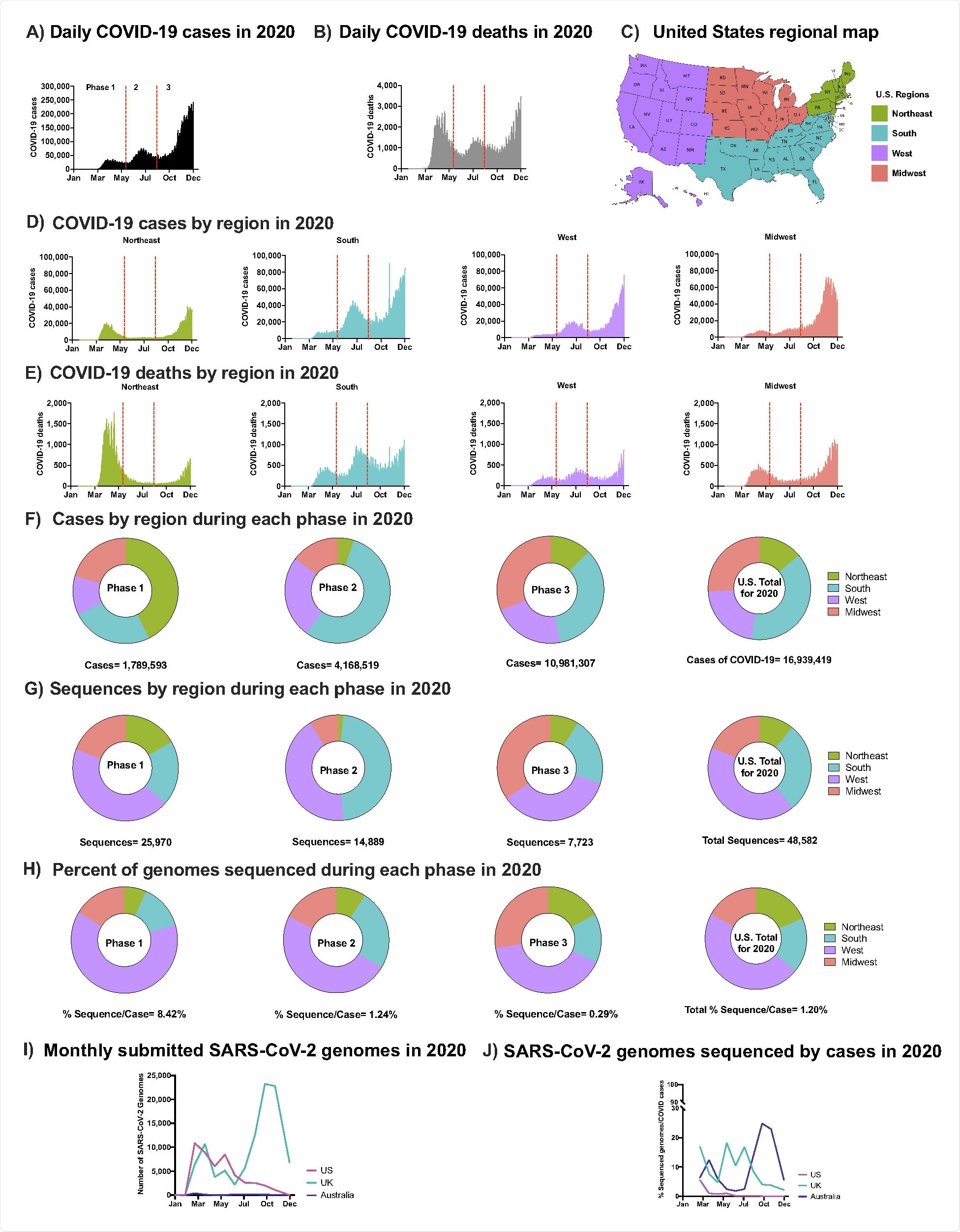
SARS-CoV-2 Epidemic in the U.S. in 2020 (A) Daily COVID-19 cases in the U.S. in 2020 (B) Daily COVID-19 deaths in the U.S. in 2020 (C) U.S. regional map colored by region (D) Number of COVID-19 cases in the U.S. in 2020 by region: Northeast, South, West, Midwest, respectively. (E) Number of COVID-19 deaths in the U.S. in 2020 by region. (A-B & D-E) Separation of Phases is denoted by vertical dotted red lines. Data were smoothed by a moving 3-day average. (F) Proportion of COVID-19 cases by region during each phase and the overall contribution to the U.S. total in 2020. (G) Proportion of SARS-CoV-2 sequences accessed (submission as of December 15th, 2020) by region during each phase and the overall contribution to the U.S. total in 2020 (H) The number of sequneces per case were obtained by each region during each phase and the U.S. total in 2020. (F-H) Highlights Phase 1, 2, and 3, followed with U.S. total of 2020. (I) Total number of sequences submitted to GISAID from the U.K., Australia, and the U.S. by December 15th, 2020. (J) Submitted SARS-CoV-2 genomes normalized to the number of COVID-19 cases from the U.K., Australia, and the U.S.
G-based clades are defined by the D614G mutation to the spike protein, being more infectious and expressing better resistance to some monoclonal antibodies than wildtype, though convalescent serum remains effective at neutralization, and clinical outcomes are similar to or even lesser than wildtype SARS-CoV-2.
Over 99% of sequences collected during phase 2 were of a G-based clade, demonstrating the rapid rise to dominance of this highly transmissible strain.
The average pair-wise distance among G-based clades rose from 0.02% in phase 1 to 0.06% in phase 3, with an approximate rate of change of 1.95 nucleotides per month.
Clades GH and GR emerged from this clade, and show even higher average mutation rates at 2.85 and 2.22 nucleotides per month, respectively.
In total, there was an increase of 14% in the number of unique variants of the G-clade throughout 2020 and an increase of 17% in the GR clade specifically.
Interestingly, the GH clade had an 11% decrease in the number of variants while the difference between variants increased.
The measure of the degree to which random populations of the virus remain non-divergent over time was also calculated for each clade, finding that G and S-based clades diverged heavily during phases 1 and 2, suggesting that viral evolution was directional. Had a great deal of unstructured mixing of differing clades taken place, divergence would be lesser, demonstrating that SARS-CoV-2 had fully pervaded the human population.
The authors state that around half of new mutations arising in the US and persisting at a frequency of over 5% were unique. The clade G nucleocapsid mutation S194L and clade GH mutations L3352F, N1653D and R2613C to ORF1a and ORF1b, respectively, increased drastically by more than 40% in representation from phase 1 to 3, and phase 3 saw the most unique mutations overall, even given the smaller available sample pool. Many of the defining mutations of SARS-CoV-2 variants of concern were identified by the group throughout 2020 before they had officially been recognized as distinct lineages. These mutations were present at a frequency of only around 1% in phase 1, rising to almost 5% in phase 3.
Future SARS-CoV-2 evolution
While SARS-CoV-2 demonstrates high replication fidelity compared with many other RNA viruses, the wide global spread of the virus has allowed ample opportunity for mutation.
The authors characterize the evolution of SARS-CoV-2 as slow but inexorable, being mainly driven by genetic drift, with some mild selection pressures towards high transmissibility and immune escape by competition with other strains.
Chronically infected immunosuppressed individuals that receive treatment with neutralizing antibodies are thought to be an ideal environment for more significant mutations to occur, proving an isolated container with more intense selection pressures, and many of the more concerning variants may have come about in this way.
Similarly, the general genetic diversity in SARS-CoV-2 has been promoted by low adherence to non-pharmaceutical measures in the community, with some adhering populations providing isolated conditions suitable for mutation before then being spread by the non-adherent.
As more of the community is vaccinated, selective pressure towards strains that better escape immune capture will be promoted. Thus continuous monitoring of the genome of the virus is essential.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
2020 SARS-CoV-2 diversification in the United States: Establishing a pre-vaccination baseline, Adam A. Capoferri, Wei Shao, Jon Spindler, John M. Coffin, Jason W. Rausch, Mary F. Kearney, medRxiv, 2021.06.01.21258185; doi: https://doi.org/10.1101/2021.06.01.21258185, https://www.medrxiv.org/content/10.1101/2021.06.01.21258185v1
- Peer reviewed and published scientific report.
Capoferri, Adam A., Wei Shao, Jon Spindler, John M. Coffin, Jason W. Rausch, and Mary F. Kearney. 2022. “A Pre-Vaccination Baseline of SARS-CoV-2 Genetic Surveillance and Diversity in the United States.” Viruses 14 (1): 104. https://doi.org/10.3390/v14010104. https://www.mdpi.com/1999-4915/14/1/104.