A team of scientists from the National Institutes of Health (NIH), USA, has recently identified six novel small molecule inhibitors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants. The study is currently available on the bioRxiv* preprint server.
Since its emergence in late December 2019 in China, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has undergone more than 12,000 mutations. The majority of these mutations have appeared in the spike protein of SARS-CoV-2, leading to the emergence of viral variants with significantly increased infectivity and immune evasion potency.
According to the Centers for Disease Control and Prevention (CDC) report, a total of 13 variants of SARS-CoV-2 have emerged globally; of which five have been designated as Variants of Concern (VOCs).
At the initial phase of the pandemic, the National Center for Advancing Translational Sciences (NCATS), USA, started screening several antiviral medicines to identify potential repurposed anti-COVID-19 therapeutics. They created the OpenData portal to make drug-related information publicly available.
Recently, scientists from the NCATS, NIH, conducted a hybrid virtual screening of two libraries of more than 120k compounds to identify potential anti-SARS-CoV-2 inhibitors with limited cytotoxicity.
The hybrid in silico approach includes quantitative structure-function relationship and ligand-based pharmacophore modeling. The scientists further evaluated the identified compounds for anti-SARS-CoV-2 activity using cytopathic effect and cytotoxicity assays.
Screening of compounds
The scientists conducted two rounds of in silico screening. From the 1st and 2nd rounds of screening, they identified a total of 640 compounds.
By analyzing SARS-CoV-2 cytopathic effect inhibiting activity and toxicity profile, they further selected 42 and 58 compounds from the 1st and 2nd rounds of screening, respectively, which showed significant antiviral activity and minimal or no cytotoxicity.
Structure-function relationships of selected compounds
The scientists identified three potential chemotypes (A, B, and C) with three or more active structural analogs by conducting hierarchical cluster analysis.
Within chemotype A, three analogs were identified with more than 83% efficacy. Within chemotype B, eight potential analogs were identified with efficacy ranging from 66% to 100%. Within quinazoline-containing chemotype C, ten potential analogs were identified, including three with the highest antiviral activity.
All analogs belonging to the three chemotypes showed acceptable solubility and permeability. However, the analogs exhibited a poor metabolic half-life. To obtain a halt-life of more than 30 min, the scientists added an N-aminoethyl group of the piperazine ring.
Mode of action of selected compounds
The scientists further evaluated the compounds identified in the hybrid screening for their activity against viral entry and replication.
Since inhibition of the SARS-CoV-2 main protease impairs the functional maturation of viral proteins, the scientists initially screened the active compounds in the SARS-CoV-2 main protease enzymatic assay. However, they could not detect any activity.
Furthermore, they tested the ability of these compounds in preventing the interaction between SARS-CoV-2 spike receptor-binding domain (RBD) and human angiotensin-converting enzyme 2 (ACE2). However, they failed to obtain conclusive results.
To overcome these shortcomings, they conducted microscale thermophoresis to test whether the compounds bind to ACE2. Moreover, they conducted a SARS-CoV-2 pseudoparticle entry assay to investigate if the compounds can interfere with SARS-CoV-2 host cell entry by binding to the viral receptor ACE2. From these analyses, they identified six compounds that bind to ACE2 with high affinity and subsequently prevent viral entry into host cells.
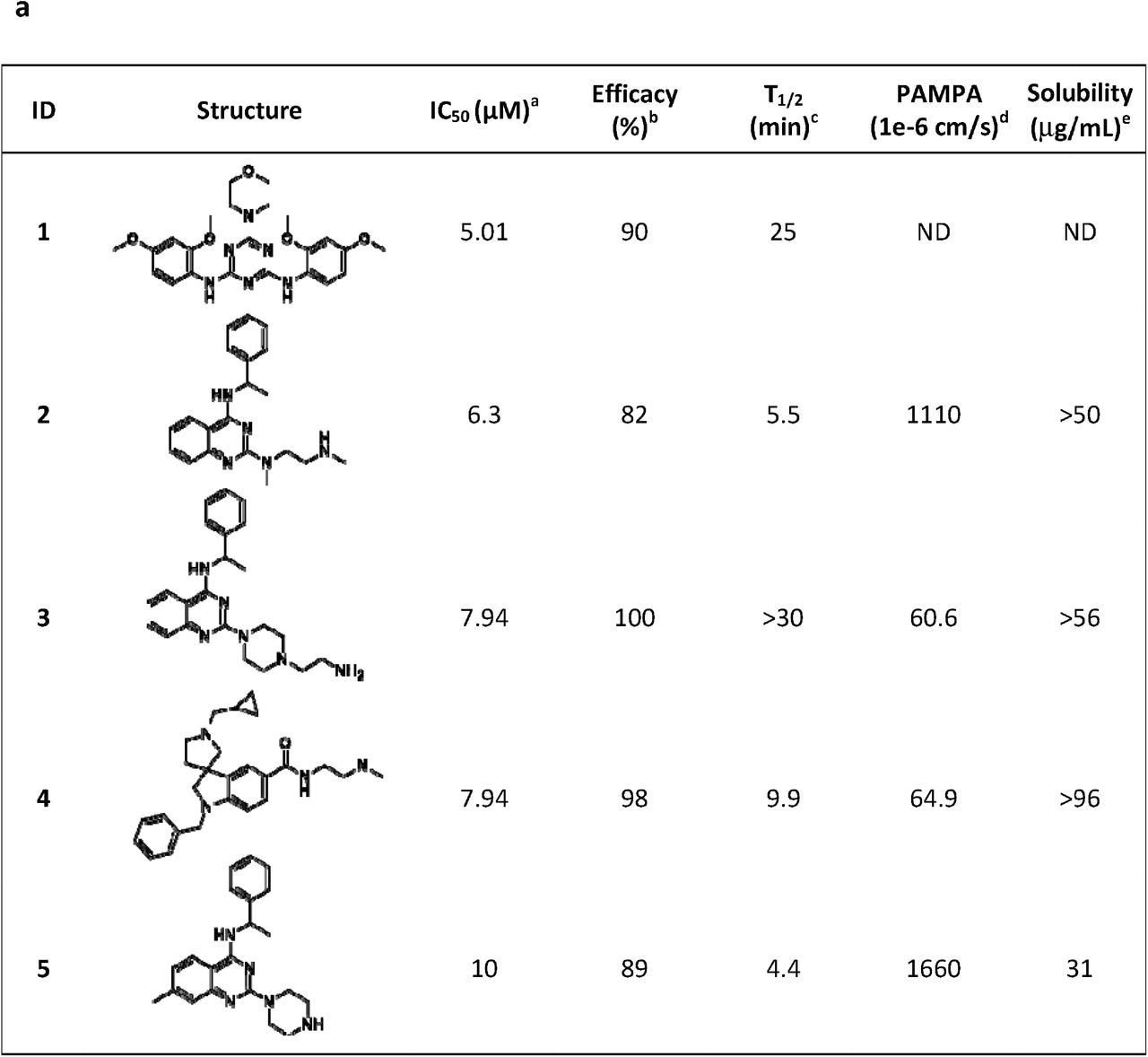
Five most potent and efficacious compounds identified, along with in vitro/physico-chemical ADME data. aIC50: half-maximal inhibitory concentration value obtained from the CPE assay in 8-point dose-response, measured in duplicate. b. Efficacy: maximum inhibitory effect observed in CPE assay. cT1/2: metabolic half-life measured in rat liver microsome lysates reported in minutes, with a minimum detectable half-life of 1 minute. dPAMPA (parallel artificial membrane permeation assay) is reported as a metric of the passive permeability of the compounds (1×10−6 cm/s). e Solubility – pION μSOL assay for kinetic aqueous solubility determination, pH 7.4. b. Three chemotypes (A-C) identified as active in the CPE assay.
Since none of the compounds showed a binding affinity for the viral spike protein, the scientists hypothesized that the antiviral activity of the compounds does not depend on the spike sequence. Thus, the compounds are expected to be effective against newly emerged viral variants with multiple spike mutations.
.jpg)
Dose-response curves of the six ACE2 binding compounds in PP and cytotoxicity assays. a, compound 1; b, compound 2; c, compound 5; d, compound 24; e, compound 25; f, compound 19. WT – wild type SARS-CoV-2 variant assay; SA – South African B.1.351 variant assay; UK – UK B.1.1.7 variant assay; VSV-G – PP assay containing the G protein of vesicular stomatitis virus; Tox – cytotoxicity assay.
To prove the hypothesis, they tested the compounds against different viral variants. They observed that the compounds have comparable or higher antiviral efficacy against the B.1.351 (first detected in South Africa) and B.1.1.7 (first detected in the UK) variants compared to that against the wildtype SARS-CoV-2.
Study significance
The study identifies six novel antiviral compounds that significantly inhibit SARS-CoV-2 host cell entry by binding to ACE2. Since the compounds do not alter the ACE2 enzymatic activity, the scientists suggest that the compounds allosterically bind to ACE2.
Importantly, the compounds are unlikely to lose their antiviral efficacy due to spike mutations as they do not directly interact with SARS-CoV-2 spike proteins.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources