Based on computational analysis, researchers have discovered multiple drugs that target specific genes and pathways altered in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections and had the potential to be potential treatments.
Although the COVID-19 has been raging around the globe for more than a year, there are have been few drugs found that are effective against the disease.
Trials have shown that Remdesivir, a drug approved by the United States FDA for COVID-19, has little effect on the death rate in hospitalized patients. Therefore, the identification of new medical therapies for both prevention and treatment is a key priority.
Repurposing drugs already approved to treat other diseases can significantly cut down the time to develop new treatments for the disease. The RNA sequencing (RNA-Seq) of lung epithelial cells infected with the SARS-CoV-2 virus can be used to analyze the relationship between gene expression triggered by infection and the progression of the infection.
Analyzing differentially expressed genes from RNA-Seq can help us better understand the infection process and find drugs that can be repurposed.
In a new study posted to the medRxiv* preprint server, researchers from the University of Pittsburgh and Barrow Neurological Institute report the results of their analysis of several RNA-Seq studies and used computational approaches to discover potential drugs against COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Computational analysis
Researchers searched the CLC Genomics Workbench database for RNA-Seq data for SARS-CoV-2. Three studies were selected, and the sequences obtained from them were mapped to the human genome. The genes obtained from this mapping were analyzed for diseases, and molecular and cellular functions.
They next obtained drug information and mechanism of action from a commercial drug database and when used in combination with all the known genes involved in COVID-19, to come up with a list of possible drug candidates.
Their analysis revealed 283 differentially expressed genes and pathway analysis identified 27 diseases and disorders, 21 cellular and molecular functions, and 275 general pathways. Infection caused by SARS-CoV-2 compromised mainly infected cells' movement, cell death/survival, as well as signaling. The genes could not only be targets for therapeutics but may also act as biomarkers for disease progression and response to treatment.
The analysis indicated the inflammatory nature of host response in respiratory cells that could lead to cell death and other respiratory problems, similar to what is seen clinically.
Based on the differentially expressed genes, a network algorithm created 25 connection networks between molecules. The most relevant network was associated with 23 genes in the dataset that were related to cellular movement and immune cell trafficking. Other main networks were associated with genes related to cellular development and growth, cell death and survival.
Potential drugs
Fifteen differentially expressed genes were related to 46 drugs, either approved by the FDA or under clinical trial. Three of these are under clinical trial for COVID-19. They also found several other drugs targeting four new genes, with six drugs that could potentially be repurposed, but have not been studied for treating COVID-19 as yet. Another eight gene targets are associated with COVID-19, but there are no drugs available that target them.
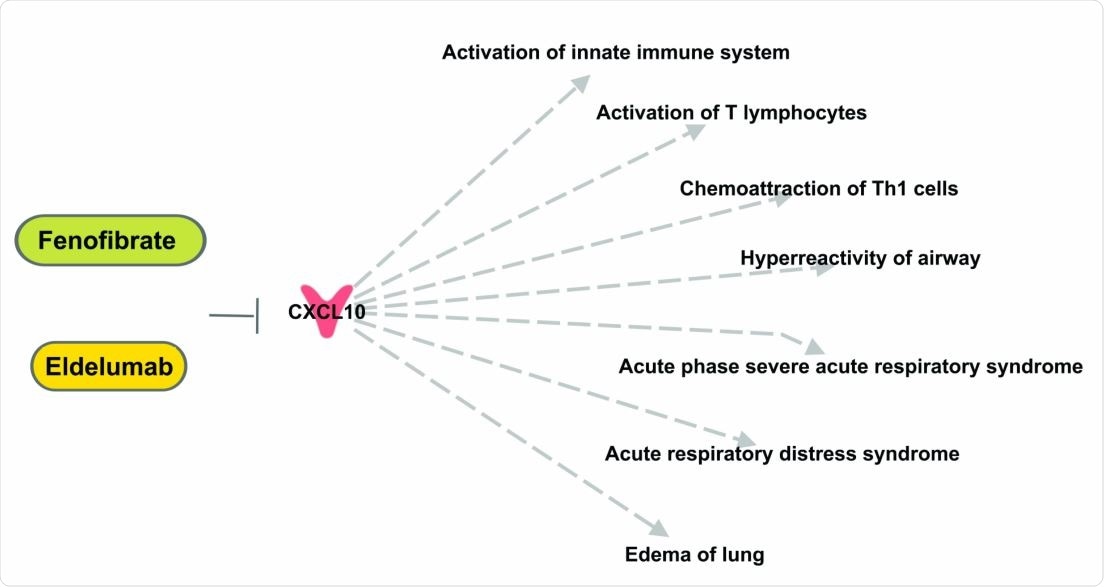
Top anti-SARS-CoV-2 target, CXCL10, affect inflammatory and immune responses. Eldelumab and Fenofibrate may be assessed for their effects in modulating COVID-19-related lung pathology.
Pathway analysis revealed activation of cytokines and an increase in cell death factors. The IL7 pathways, which regulate important inflammatory cytokines, were also enriched. Although there is growing evidence of the role of IL7 in COVID-19 progression, further studies need to be performed to understand how inhibiting it may help.
Using online gene ranking algorithms, the team also ranked the genes to prioritize targets against SARS-CoV-2. The topmost ranked gene was CXCL10, which is involved in the regulation of inflammatory and immune responses, and could be a potential target for drugs to treat COVID-19.
CXCL10 has been associated with other viral infections like SARS-CoV, Ebola, and dengue. Increased levels are positively associated with organ damage. Eldelumab and fenofibrate are drugs that may decrease CXCL10 activity, which could be tested further for COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources