A recent tour-de-force study by researchers from the United Kingdom reveals variants of the human angiotensin-converting enzyme 2 (ACE2) that can improve or decrease binding of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – including two variants with distinct population distributions showing an enhanced affinity for viral spike glycoprotein. This research paper is currently available on the bioRxiv* preprint server while the article undergoes peer review.
Since the start of the coronavirus disease 2019 (COVID-19) pandemic, caused by the SARS-CoV-2, one of the most pertinent questions was why there is such a diverse array of disease outcomes. More specifically, besides older age and the presence of comorbidities, it was not clear whether a genetic component may also play a role in predisposing individuals to a dire prognosis.
And while different genetic association studies have already pinpointed several loci implicated in COVID-19 risk, further steps towards elucidating genetic factors of COVID-19 susceptibility could have important implications not only for clinical decision making but also for epidemic dynamics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The importance of missense mutations
It was previously shown that missense variants observed in protein-protein interaction interfaces could give rise to altered binding characteristics and affect susceptibility in the context of virus-host interactions. This finding implies the potential of missense variants in ACE2 to influence a key step in SARS-CoV-2 infection, which is a receptor-binding event.
Missense mutations basically arise when there is a change of a single base pair, resulting in different amino acids in the protein, exhibiting further downstream effects. A novel computational prediction algorithm, mCSM-PPI2, was recently developed to predict the effects of missense mutations on interaction binding affinity.
This was utilized by Professor Geoffrey J. Barton and Dr. Stuart A. MacGowan from the Division of Computational Biology, School of Life Sciences of the University of Dundee, who applied the aforementioned algorithm to assess the effects of cell receptor genetic variability on the binding of SARS-CoV-2 spike glycoprotein and revealed specific ACE2 variants.
Now, a more extensive research group from the UK (also led by Senior Bioinformatics Research Scientist Dr. Stuart A. MacGowan) went one step further and aimed to appraise binding affinities of 10 ACE2 variants with receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein.
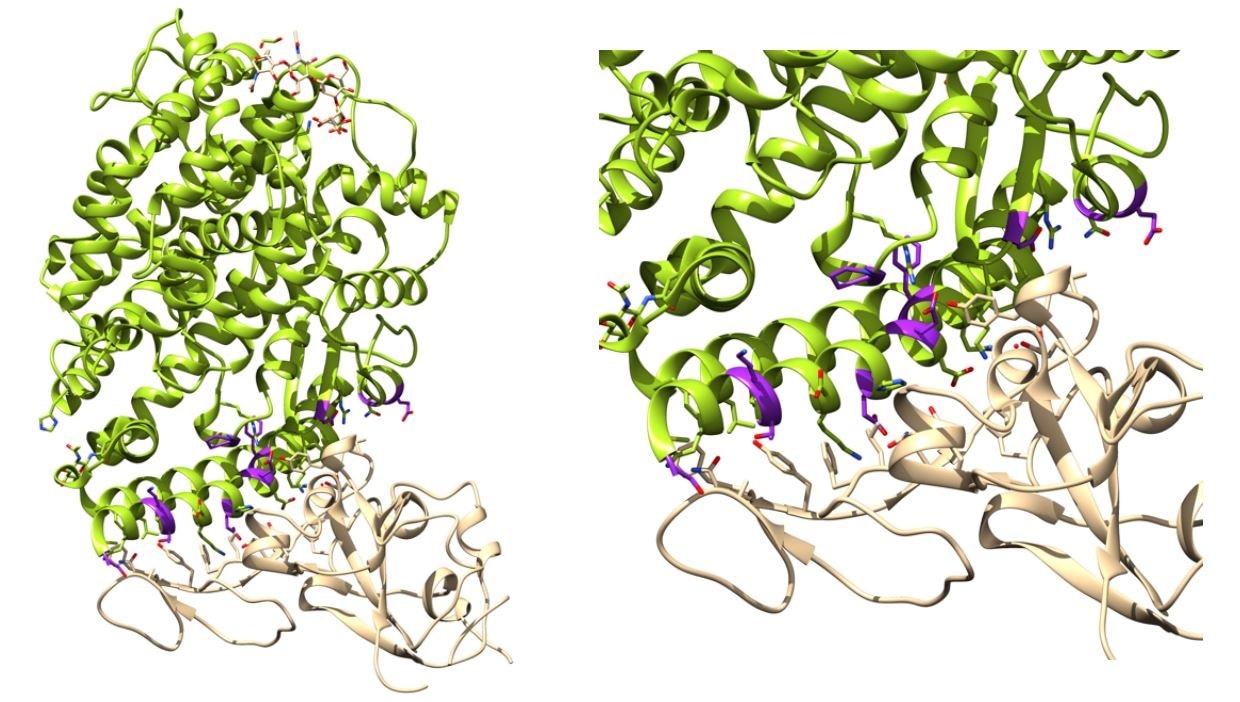
Binding affinity determination of ACE2 variants with SARS-CoV-2 Spike. Left: ACE2 (green) in complex with Spike RBD (tan) complex from biological assembly 1 derived from PDB ID: 6vw111. The positions that were mutated in this work are highlighted magenta. Right. The ACE2 Spike interface. Figure generated with Jalview and UCSF Chimera.
Exploring the Genome Aggregation Database
In short, the researchers selected variants based on affinity predictions and prevalence in the Genome Aggregation Database (gnomAD), which is a library of exome and genome sequencing data from a myriad of large-scale sequencing projects. Their affinities for RBD of spike glycoprotein have been measured via surface plasmon resonance (SPR).
From the 241 ACE2 missense variants reported in gnomAD, nine (out of ten) mutants were selected based on the authors’ previous computational predictions and their reported prevalence. The tenth mutant was predicted to enhance spike glycoprotein binding more than any other possible mutation at the interface.
Such an approach was pursued to obtain better insight into the effect of ACE2 variants on SARS-CoV-2 spike glycoprotein binding and reveal additional variants that may enhance this process. Moreover, the SPR data allowed the researchers to recalibrate previous mCSM-PPI2 predictions and yield much more accurate estimates of the effect of interface variants.
The structure of ACE2 (green) gnomAD24 missense variant p.Ser19Pro enhances Spike (light blue) binding affinity. A. The environment of ACE2 Ser19 from PDB ID: 6vw111. B. Model of ACE2 p.Ser19Pro in complex with Spike. The mutant structure was modeled onto 6vw1 with mCSM-PPI13.
Variants that can enhance or reduce binding
This study found ACE2 variants that could either enhance or reduce binding, including two variants with distinct population distributions that enhanced affinity for spike glycoprotein. The enhancers and, thus, possible risk factors for COVID-19 were ACE2 p.Ser19Pro (predominant in gnomAD African cohort) and p.Lys26Arg (pervasive in the Ashkenazi Jewish cohort).
On the other hand, three rare ACE2 variants that firmly inhibited or abolished spike glycoprotein binding were ACE2 p.Glu37Lys, p.Gly352Val and p.Asp355Asn. Consequently, the presence of these variants may confer resistance to infection with SARS-CoV-2.
Albeit ACE2 p.Glu37Lys and p.Asp355Asn were already predicted as inhibitors of spike binding before and potentially protective against COVID-19, two additional rare variants were also identified in this study. This includes p.Gly326Glu, which was previously erroneously predicted as a binding enhancer.
Predictive diagnostics and population risk assessment
Pinpointing specific genetic variants with the potential to influence the severity and course of COVID-19 provides a unique opportunity for predictive diagnostic approaches, intervention in the early stages of the disease and personalized treatment modalities, but also for population risk assessment.
“A key point in these burden assessments is the separate consideration of variants predicted to inhibit and enhance spike affinity since these may have distinct phenotypes,” say the authors of this bioRxiv paper.
“Overall, p.Ser19Pro and p.Lys26Arg are still expected to have the most widespread effects, with the joint prevalence of the rare affinity variants substantially lower, but in all cases, the possibility of higher prevalence in local or underrepresented populations remains and the penetrance of each variant will be a key factor”, they add.
In conclusion, this work provides many significant insights for further experimental work into viral recognition of human ACE2 receptors, but also for understanding SARS-CoV-2 spike glycoprotein evolution and host adaptation. However, even real-world clinical and epidemiological usage may stem from such meticulous predictive research endeavors.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references: