The emergence of newer variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), continues to pose new threats to public health. These variants, many of which have been referred to as variants of concern (VOCs), are often more transmissible and/or more virulent than the wild-type lineage.
This is an important finding, as the transmission risk of a virus is determined not only by its genotype but also by other competing lineages that are in circulation at the time, as well as the level of antiviral immunity present in the local population.
Background
The B.1.1.7 and B.1.526 SARS-CoV-2 variants are also known as the Alpha and Iota variants, respectively. Whereas the B.1.1.7 variant was originally identified in the United Kingdom, the second was first identified in New York, both towards the end of 2020.
The Alpha variant quickly became the dominant strain circulating throughout the United Kingdom shortly after its identification. Once the Alpha variant reached the United States, its increased transmissibility subsequently allowed for it to also become the dominant strain in this nation as well.
Notably, the co-circulation of the Iota variant throughout New York at the same time that the B.1.1.7 variant was in circulation may have slowed the incidence of COVID-19 in this area of the United States.
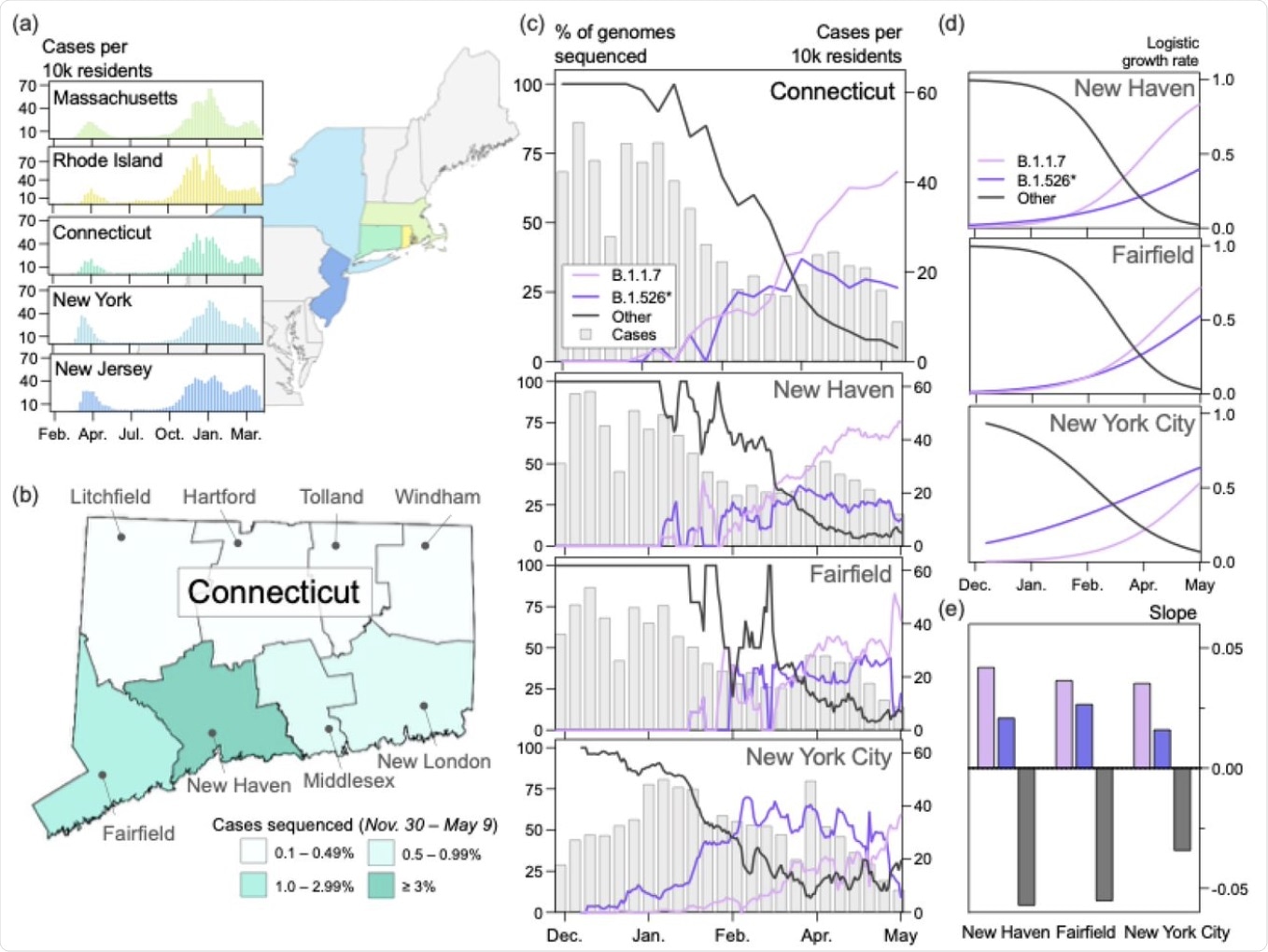 B.1.1.7 and B.1.526* dominated the circulating SARS-CoV-2 populations in Connecticut and New York City in early 2021. (a) Trends in COVID-19 incidence were consistent across northeastern states throughout the pandemic. (map) Connecticut (teal) is bordered by New York, Rhode Island, and Massachusetts. New York City is less than 50 miles from Fairfield County. Weekly COVID-19 incidence was tabulated according to the Johns Hopkins COVID-19 portal (https://github.com/CSSEGISandData/COVID-19). Shapefile source: United States Census Bureau. (b) New Haven County led the state in the percentage of COVID-19 cases sequenced between November 30, 2020 and May 9, 2021 (3.33%). During this period, 0.51% of COVID-19 cases in New York City were sequenced. Genomes that were collected through targeted variant screening (e.g., spike-gene target failure) were excluded from this analysis. Shapefile source: the Connecticut Department of Energy & Environmental Protection (DEEP) Geographic Information Systems Open Data Website. (c)Together, B.1.1.7 and B.1.526* variants displaced nearly all other SARS-CoV-2 lineages in New Haven County (n = 2,086), Fairfield County (n = 612), and New York City (n = 4,528). The lineages of sequenced viruses were assigned using pangolin v.2.4.2. The lineages B.1.526, B.1.526.1, and B.1.526.2 were assigned to the general lineage category ‘B.1.526*’. We calculated a 7-day rolling average for the proportion of B.1.1.7, B.1526*, and ‘other’ SARS-CoV-2 lineages sequenced in our dataset. (d) Logistic regression of the growth rates per lineage using Rv.4.0.1. Line colors correspond to the legend in (c). (e) Slope of logistic growth shown in (d). Bar colors correspond to the legend in (c).
B.1.1.7 and B.1.526* dominated the circulating SARS-CoV-2 populations in Connecticut and New York City in early 2021. (a) Trends in COVID-19 incidence were consistent across northeastern states throughout the pandemic. (map) Connecticut (teal) is bordered by New York, Rhode Island, and Massachusetts. New York City is less than 50 miles from Fairfield County. Weekly COVID-19 incidence was tabulated according to the Johns Hopkins COVID-19 portal (https://github.com/CSSEGISandData/COVID-19). Shapefile source: United States Census Bureau. (b) New Haven County led the state in the percentage of COVID-19 cases sequenced between November 30, 2020 and May 9, 2021 (3.33%). During this period, 0.51% of COVID-19 cases in New York City were sequenced. Genomes that were collected through targeted variant screening (e.g., spike-gene target failure) were excluded from this analysis. Shapefile source: the Connecticut Department of Energy & Environmental Protection (DEEP) Geographic Information Systems Open Data Website. (c)Together, B.1.1.7 and B.1.526* variants displaced nearly all other SARS-CoV-2 lineages in New Haven County (n = 2,086), Fairfield County (n = 612), and New York City (n = 4,528). The lineages of sequenced viruses were assigned using pangolin v.2.4.2. The lineages B.1.526, B.1.526.1, and B.1.526.2 were assigned to the general lineage category ‘B.1.526*’. We calculated a 7-day rolling average for the proportion of B.1.1.7, B.1526*, and ‘other’ SARS-CoV-2 lineages sequenced in our dataset. (d) Logistic regression of the growth rates per lineage using Rv.4.0.1. Line colors correspond to the legend in (c). (e) Slope of logistic growth shown in (d). Bar colors correspond to the legend in (c).
How was the study done?
The current study used a different framework to directly compare the transmissibility of these variants, in which both epidemiological and genomic data were collected and analyzed from Connecticut between January 2021 to May 2021. Assessing the spread rate from the rate of growth and the effective reproduction number (Rt) as it changed over time, the scientists concluded that the Alpha variant was 6-10% more transmissible than the Iota variant when both were circulating in the same population.
This finding corroborated earlier observations made in New York City, where the Iota variant was already circulating before the introduction of the Alpha variant. These variants replaced almost all other strains already in circulation within three months. However, the Alpha variant accounted for a larger proportion of new COVID-19 cases during this period.
Secondly, the researchers of the current study explored the introductions of each variant into Connecticut by clade size, timing, and number. This ruled out the influence of higher introductions as an explanation of the larger clade size of the Alpha variant and instead attributed its rising presence to higher viral fitness.
What were the findings?
Both the Alpha and Iota variants were found to be circulating throughout Connecticut in the early months of 2021 as a result of infected travelers visiting this state. Instead of one strain becoming dominant over the other, both variants circulated simultaneously throughout Connecticut.
While both the Alpha and Iota variants were circulating throughout this area of the U.S. at the same time, the Alpha variant was growing at double the rate of the Iota variant both in New York and in New Haven County in Connecticut. Comparably, in the neighboring county of Fairfield in Connecticut, the Alpha variant showed a 37% higher growth rate as compared to the Iota variant.
The same framework was then used to analyze thousands of randomly sampled viral genomes obtained from surveillance data in New York City. This yielded the same trends in the transmissibility of the two variants.
The second phase of the study was based on the more traditional phylogeographic analysis. This showed that the Alpha variant came from both international and domestic travelers, as it was widespread in the both the United Kingdom and Europe.
In contrast, the Iota variant was found to spread from New York City into Connecticut. Despite a marginal reduction in the frequency of the Alpha variant arriving in Connecticut, larger clusters of cases were still detected in this state.
The results showed that both variants were introduced into Connecticut at approximately the same rates; however, the Alpha variant led to a much larger clade size. Thus, both threads of research indicate that the Alpha variant has greater viral fitness as compared to the Iota variant, with both showing greater fitness than earlier variants.
As vaccination rates increased, both variants showed a reduction in their Rt values; however, this value for the Alpha variant remained higher than that for the Iota variant. By the end of April 2021, which is when a quarter of eligible people had been vaccinated in Connecticut, the Rt remained above 1, showing that the outbreak was continuing to grow.
Eventually, the Rt for the Iota variant fell below 1, indicating reduced transmission one week earlier than that which was detected for the Alpha variant. Conversely, for other SARS-CoV-2 variants aside from the Alpha and Iota variants, the Rt values were suggestive of declining infections early in January 2021.
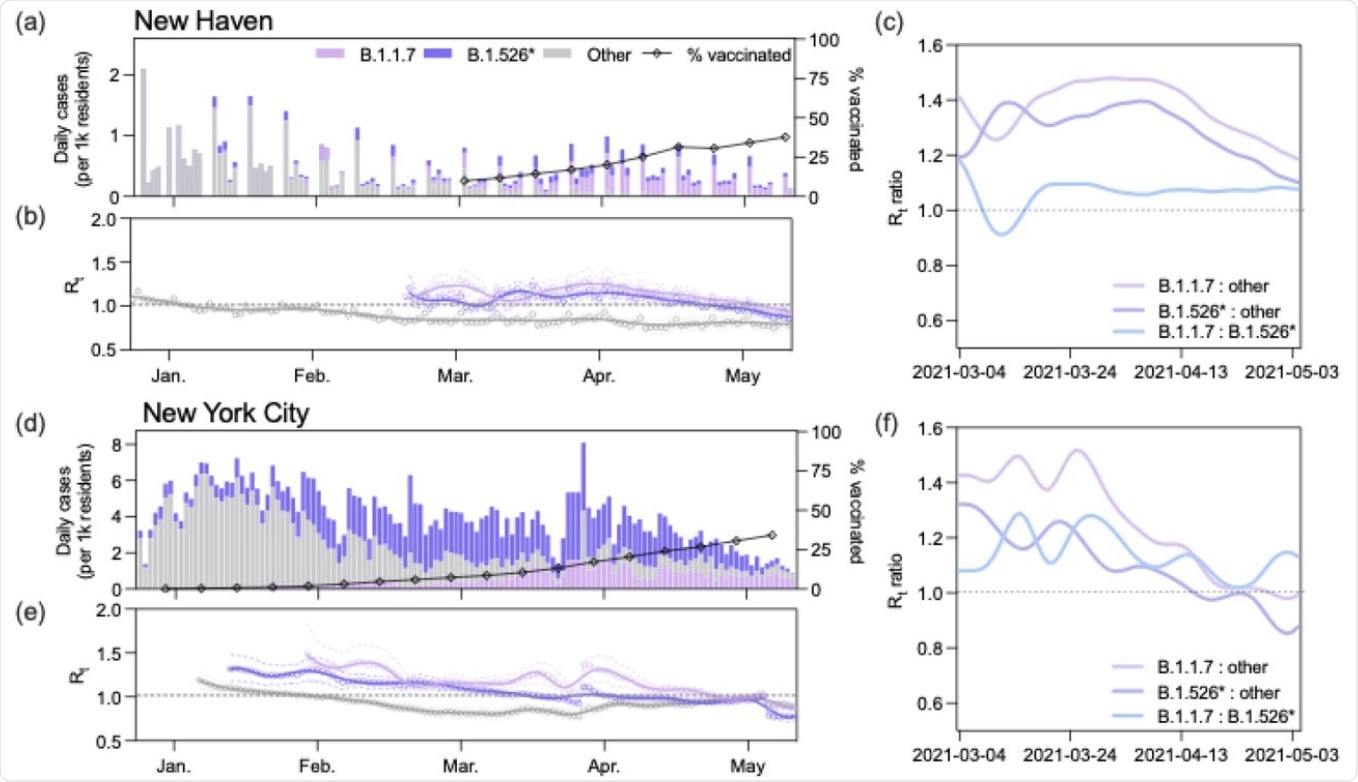 B.1.1.7 had a larger effective reproduction number (Rt) than B.1.526 during the COVID-19 vaccination campaign. (a, d) Daily incidence and full vaccination rates (2 weeks post last dose) of B.1.1.7, B.1.526*, and other circulating lineages in New Haven County (a) and New York City (d). Daily cases were assigned to one of three lineage categories (‘B.1.1.7’, ‘B.1.526*’, and ‘other’) according to the 7-day rolling average of variant frequency among sequenced cases. ‘B.1.526*’ includes the sublineages B.1.526, B.1.526.1, and B.1.526.2. (b, e) Time-varying effective reproduction numbers (Rt) were calculated using the R package epiEstim. An Rt value above 1 indicates that an infected individual will, on average, infect more than 1 additional person. We assumed a serial interval of mean 5.2 days and standard deviation of 4 days for all lineages. We used a smoothing spline to smooth the daily Rt curves (line) with the package, stat in R v4.0.1. Non-smoothed estimates are shown as individual points. 0.025 and 0.975 quantiles are shown as dotted lines (c, f) Ratios of estimated Rt between March 4 and May 4, 2021 calculated using the splines shown in (b) and (e).
B.1.1.7 had a larger effective reproduction number (Rt) than B.1.526 during the COVID-19 vaccination campaign. (a, d) Daily incidence and full vaccination rates (2 weeks post last dose) of B.1.1.7, B.1.526*, and other circulating lineages in New Haven County (a) and New York City (d). Daily cases were assigned to one of three lineage categories (‘B.1.1.7’, ‘B.1.526*’, and ‘other’) according to the 7-day rolling average of variant frequency among sequenced cases. ‘B.1.526*’ includes the sublineages B.1.526, B.1.526.1, and B.1.526.2. (b, e) Time-varying effective reproduction numbers (Rt) were calculated using the R package epiEstim. An Rt value above 1 indicates that an infected individual will, on average, infect more than 1 additional person. We assumed a serial interval of mean 5.2 days and standard deviation of 4 days for all lineages. We used a smoothing spline to smooth the daily Rt curves (line) with the package, stat in R v4.0.1. Non-smoothed estimates are shown as individual points. 0.025 and 0.975 quantiles are shown as dotted lines (c, f) Ratios of estimated Rt between March 4 and May 4, 2021 calculated using the splines shown in (b) and (e).
What are the implications?
“Our approach is more informative than the current practice of tracking variant prevalence because it accounts for both the change in lineage frequencies and the number of incident cases.”
An earlier study reported that the B.1.526 variant with the spike E484K mutation showed a more rapid growth rate than the Alpha variant. However, in the current study, this observation was seen to be short-lived, with the latter becoming the dominant variant soon afterward. The rise in the Rt value of the Alpha variant until it exceeded that of the Iota variant was observed by February of 2021.
The novel framework discussed in this study is therefore useful in monitoring the changing patterns of SARS-CoV-2 transmission over time. Since this approach is based on open-source computational methods, it is a more accessible and robust tool for this type of viral surveillance.
As the pandemic progresses, the exact contribution of each new variant will need to be rapidly assessed to ensure that effective steps can be taken to control its spread.
“A genomic surveillance infrastructure coupled with the application of our framework would enable the monitoring of variant epidemiology in close to real time. The phylodynamic methods we applied in this study can be run on a desktop computer in a few hours, making the collation of representative datasets the rate limiting step.”
The study also emphasizes the importance of understanding where novel variants arise, as this will hinder the control and prevent the transmission of the virus via measures that are focused on particular entry points. Rather than prohibiting the direct entry of specific types of travelers, strong and effective measures to prevent community transmission are more likely to improve these situations as they arise.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Petrone, M. E., Rothman, J. E., Breban, M. I., et al. (2021). Combining genomic and epidemiological data to compare the transmissibility of SARS-CoV-2 lineages. medRxiv preprint. doi:10.1101/2021.07.01.21259859. https://www.medrxiv.org/content/10.1101/2021.07.01.21259859v1
- Peer reviewed and published scientific report.
Petrone, Mary E., Jessica E. Rothman, Mallery I. Breban, Isabel M. Ott, Alexis Russell, Erica Lasek-Nesselquist, Hamada Badr, et al. 2022. “Combining Genomic and Epidemiological Data to Compare the Transmissibility of SARS-CoV-2 Variants Alpha and Iota.” Communications Biology 5 (1). https://doi.org/10.1038/s42003-022-03347-3. https://www.nature.com/articles/s42003-022-03347-3.