Multivalent protein-based minibinders hold great promise as potential prophylactic and treatment agents, as they can retain their activity against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern. This was revealed in a recent tour-de-force research study currently available on the bioRxiv* preprint server while the article undergoes the peer review process.
Escape variants of SARS-CoV-2, the causative agent of the ongoing coronavirus disease (COVID-19) pandemic, threaten our progress in defeating this enormous public health hazard. Hence, alongside vaccines that are now pervasive in many countries, we need new additions to our treatment armamentarium.
Monoclonal antibodies that target the SARS-CoV-2 spike glycoprotein can improve outcomes for patients that have COVID-19. Nonetheless, producing them in sufficient quantities for use on a population level during a global pandemic is neither technically nor financially feasible.
In addition, many monoclonal antibodies are somewhat sensitive to viral escape by conferring point mutations in their recognition epitope on the spike glycoprotein trimer. This problem was addressed by preparing a cocktail of different monoclonal antibodies targeting different epitopes (which are parts of the antigen where antibodies attach), but it was shown that new variants of concern could escape this as well.
As an alternative to monoclonal antibodies, previous work has used computational power to design miniproteins with the propensity of blocking the SARS-CoV-2 receptor-binding domain (RBD) interaction of the spike glycoprotein trimer with its host receptor angiotensin-converting enzyme 2 (ACE2). They were conveniently dubbed ‘minibinders’.
In this new methodologically astonishing paper, a large research group from the United States developed multivalent versions of the minibinders – with geometries that enable concomitant engagement of all three RBDs in a single spike glycoprotein trimer, addressing the problem of SARS-CoV-2 mutations head-on.
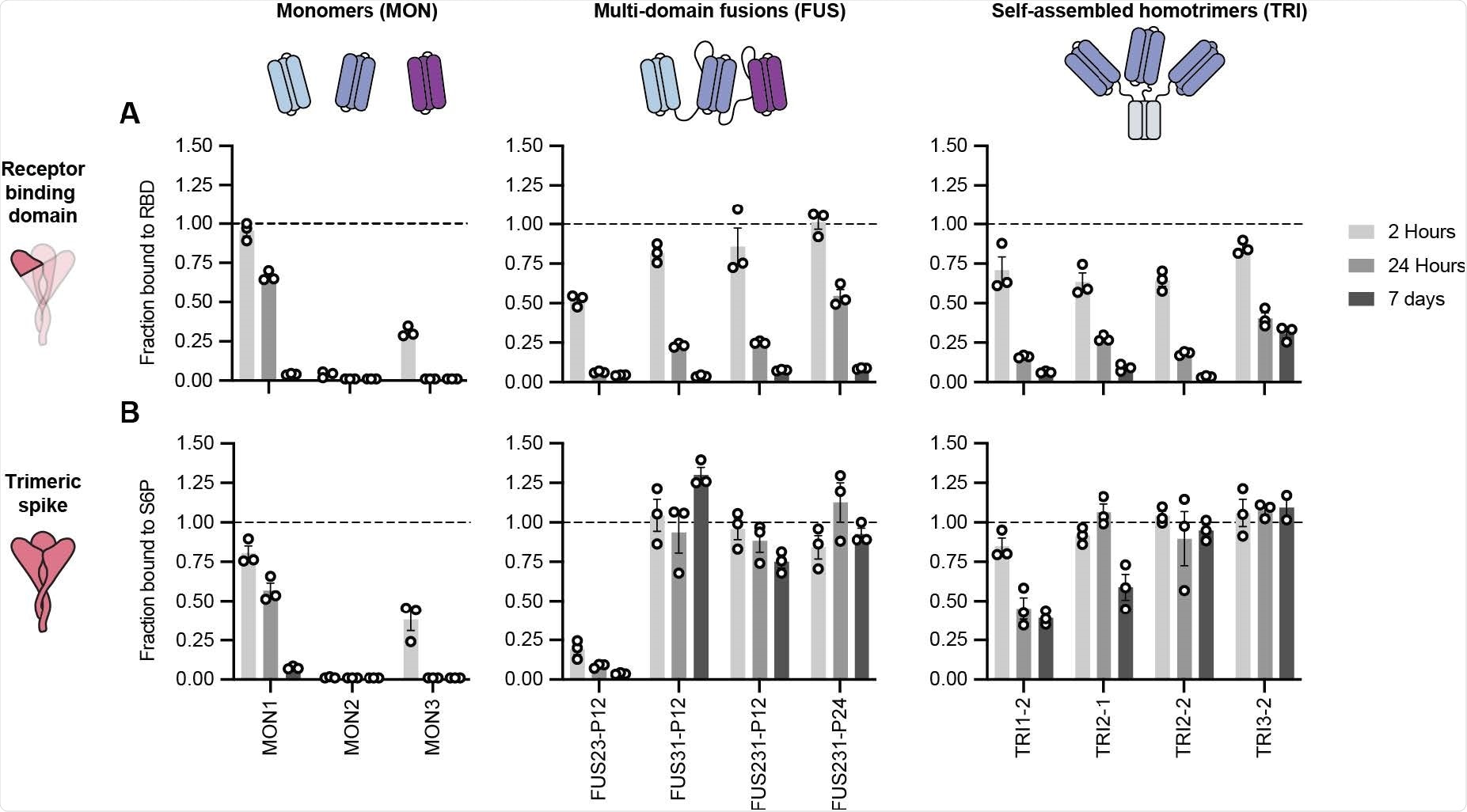
Multivalent minibinders exhibit unusually slow dissociation rates upon binding to the prefusion SARS-CoV-2-S glycoprotein. (A, B) Dissociation of the minibinder construct complexed with either the receptor binding domain (RBD) (A) or S trimer (S6P) (B) was monitored via competition with 100-fold molar excess of untagged MON1 using AlphaLISA (Mean ± SEM, n = 3 replicates from a single experiment).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Constructing and testing minibinders
In short, homotrimers of single minibinders and fusions of three distinct minibinders were constructed to geometrically match the SARS-CoV-2 spike glycoprotein trimer architecture and then further optimized by cell-free expression in order to reduce measurable dissociation upon binding.
Then, neutralization assays with the inhibitors and plaque assays were conducted with the use of both a pseudotyped lentivirus and authentic SARS-CoV-2 variants in order to investigate the efficacy of minibinders in preventing viral infection.
Finally, in order to appraise the potential of their multivalent minibinders in preventing or treating SARS-CoV-2 infection in vivo, the researchers have evaluated both pre-exposure and post-exposure therapy in highly susceptible K18-hACE2 transgenic mice.
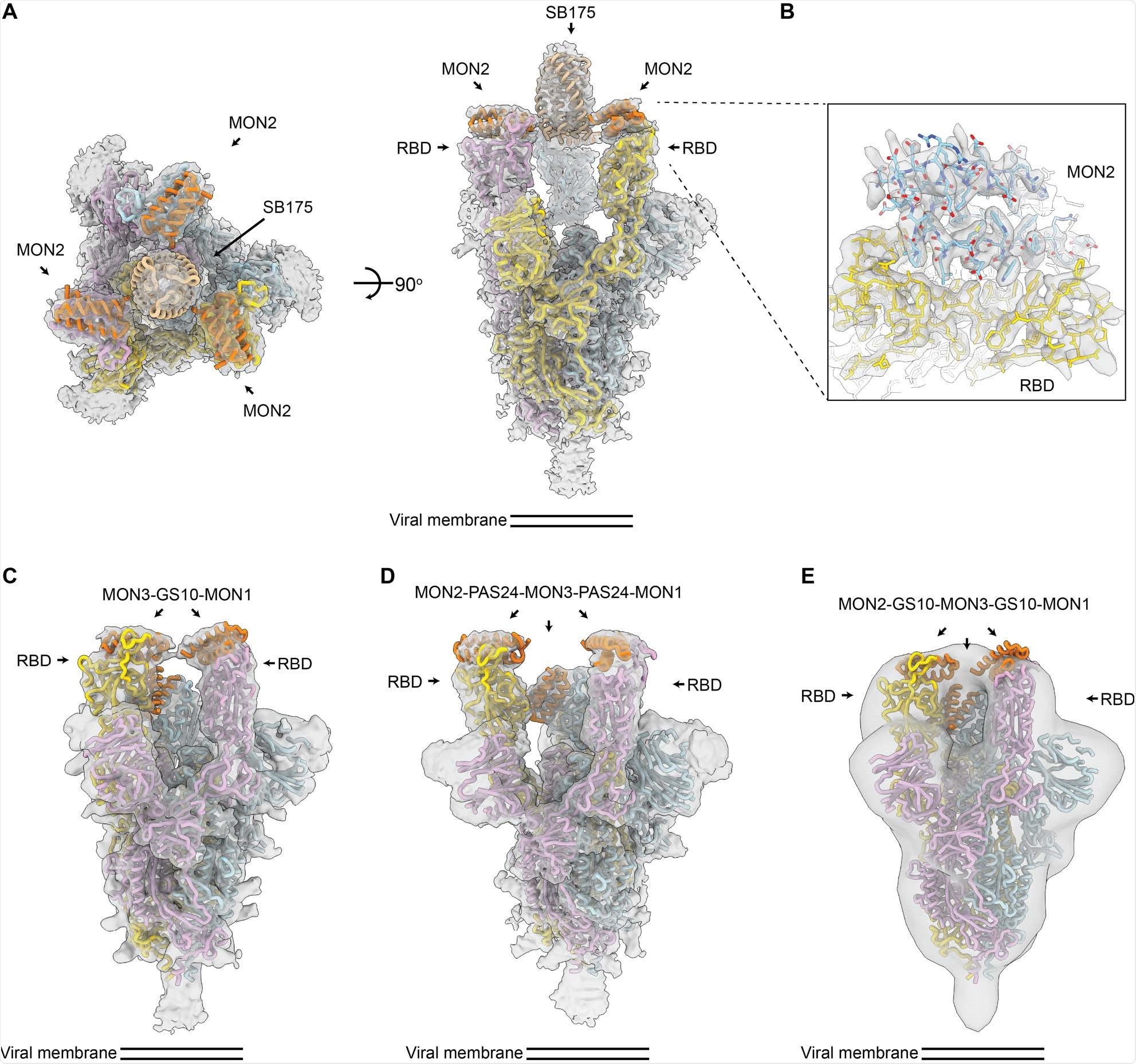
CryoEM structures of multivalent minibinders in complex with the SARS-CoV-2 S6P glycoprotein. (A) CryoEM map of TRI2-2 in complex with the S6P in two orthogonal orientations. (B) Zoomed-in view of the TRI2-2 and S6P complex interface obtained using local refinement of the RBD and TRI2-2. The RBD and MON2 built at 2.9 Å resolution are shown in yellow and blue, respectively. (C) CryoEM map of FUS31-G10 in complex with two RBDs. (D) CryoEM map of FUS231-P24 in complex with three RBDs. (E) Negative-stain EM map of FUS231-G10 in complex with S6P. The S trimer and minibinder models are placed in the whole map by rigid body fitting. EM density is shown as a transparent gray surface with a fitted atomic model. Spike protomers (PDB 7JZL) are shown in yellow, blue, and pink. Minibinders (PDB 7JZU, 7JZM, and MON2 atomic structure in this study) are shown in orange.
Bound to resist viral escape
The use of cryo-electron microscopy has demonstrated that developed trivalent minibinders can engage all three receptor binding domains on a single spike glycoprotein trimer. The results of this study have also highlighted the precision with which both protein binding interfaces and protein scaffolds can now be computationally designed.
Furthermore, the top candidates can successfully neutralize SARS-CoV-2 variants of concern with values of the half-maximal inhibitory concentration (IC50) in the low picomolar range. They can also resist viral escape and provide ample protection in highly vulnerable human ACE2-expressing transgenic mice – both as prophylactic and treatment agents.
Among them, the top candidate protein recognized was TRI2-2, shown to potently neutralize historical and current SARS-CoV-2 variants of concern. Basically, this is the first miniprotein developed to engage the spike glycoprotein in a trivalent fashion and with an intrinsically escape-resistant binding interaction.
A useful strategy for pandemic preparedness
This type of integrated computational workflow can actually boost the development of mutationally resilient therapeutics for pandemic preparedness, while the coalescence of trivalency and receptor mimicry may be a novel and valuable approach for combating escape resistance. There are also several advantages in comparison to monoclonal antibodies.
“These multivalent minibidners are amenable to large-scale and low-cost production in microorganisms like Escherichia coli, they can readily be administered directly to the respiratory system, and their stability may obviate the need for cold chain storage”, explain the authors of this study.
Looking forward, the experimental screening pipeline (together with integrated computational design) for recognizing geometrically fitting arrays of receptor mimicking minibinders may actually turn into a powerful and expeditious strategy for developing not only protein-based countermeasures but also diagnostic reagents against pathogens that may emerge in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hunt, A.C. et al. (2021). Multivalent designed proteins protect against SARS-CoV-2 variants of concern. bioRxiv. https://doi.org/10.1101/2021.07.07.451375, https://www.biorxiv.org/content/10.1101/2021.07.07.451375v1
- Peer reviewed and published scientific report.
Hunt, Andrew C., James Brett Case, Young-Jun Park, Longxing Cao, Kejia Wu, Alexandra C. Walls, Zhuoming Liu, et al. 2022. “Multivalent Designed Proteins Neutralize SARS-CoV-2 Variants of Concern and Confer Protection against Infection in Mice.” Science Translational Medicine 14 (646). https://doi.org/10.1126/scitranslmed.abn1252. https://www.science.org/doi/10.1126/scitranslmed.abn1252.
Article Revisions
- Apr 11 2023 - The preprint preliminary research paper that this article was based upon was accepted for publication in a peer-reviewed Scientific Journal. This article was edited accordingly to include a link to the final peer-reviewed paper, now shown in the sources section.