Numerous reports indicate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spillovers have occurred from humans to animals since the COVID-19 pandemic began, as evidenced by the transmission of the virus between keepers and tigers and lions at the New York Bronx Zoo.
To date, however, it is unclear what species of animals are susceptible to SARS-CoV-2. This type of information can usually be gathered by infecting various animal species with a virus to determine if they are susceptible. However, in order to reduce animal testing and refine animal experiments, researchers set out to answer this question in a more elegant and animal-friendly way.
A team of researchers at the Institute of Infectious Diseases (IFIK) at the University of Bern and the Institute of Virology and Immunology (IVI) has utilized a unique collection of advanced cell culture models of cells lining the airways from various domesticated and wildlife animals. This way, the team can determine which animals are susceptible to SARS-CoV-2 infection.
In the study, published in the U.S. Centers for Disease Control and Prevention (CDC)’s Emerging Infectious Diseases journal, the team revealed that SARS-CoV-2 efficiently infected the respiratory cells from cats and monkeys, suggesting that infection surveillance is important for these animals.
SARS-CoV-2 in animals
Over the past two decades, scientists have observed zoonotic outbreaks of severe acute respiratory syndrome (SARS) in China in 2003 and the Middle East respiratory syndrome (MERS) in Saudi Arabia in 2012. The outbreaks have been followed by the current COVID-19 pandemic, which has caused over 191 million cases worldwide.
Humans appear to be the main hosts of SARS-Cov-2, but its zoonotic origin and intermediate and spillover hosts are unknown. Studies in the past have shown that these spillover events can happen from humans to animals.
Scientists believe that close human-to-animal interactions drive zoonotic events. Throughout the pandemic, several reports of animals being infected with SARS-CoV-2 have emerged. These include infection in pets like dogs and cats, and zoo animals like big cats (lion, tiger, cougar, leopard, and puma), and gorillas. Farm animals like minks were reported to have contracted the virus, too.
In studies, animal models are essential to study the host spectrum. Aside from that, these studies can shed light on the viral pathogenesis of SARS-CoV-2, testing novel antiviral drugs, immunotherapies, and vaccines.
The study
In the current study, the researchers evaluated the susceptibility of various mammal species to SARS-CoV-2 by recapitulating the initial stages of infection in a controlled in vitro model.
The team used the repository of well-differentiated airway epithelial cell (AEC) cultures from various domesticated and wildlife animal species to arrive at the study findings. In total, cell cultures from 12 mammal species were examined, including rhesus macaque, cat, ferret, dog, pig, rabbit, goat, cattle, llama, camel, and two neotropical bat species.
The cells were isolated from deceased animals, and these cells can be increased in a petri dish. Hence, the researchers do not need to perform any animal experiments. To evaluated the susceptibility of the animal species to SARS-CoV-2, the 12 well-differentiated mammal ACE culture models were infected and monitored for viral replication kinetics.
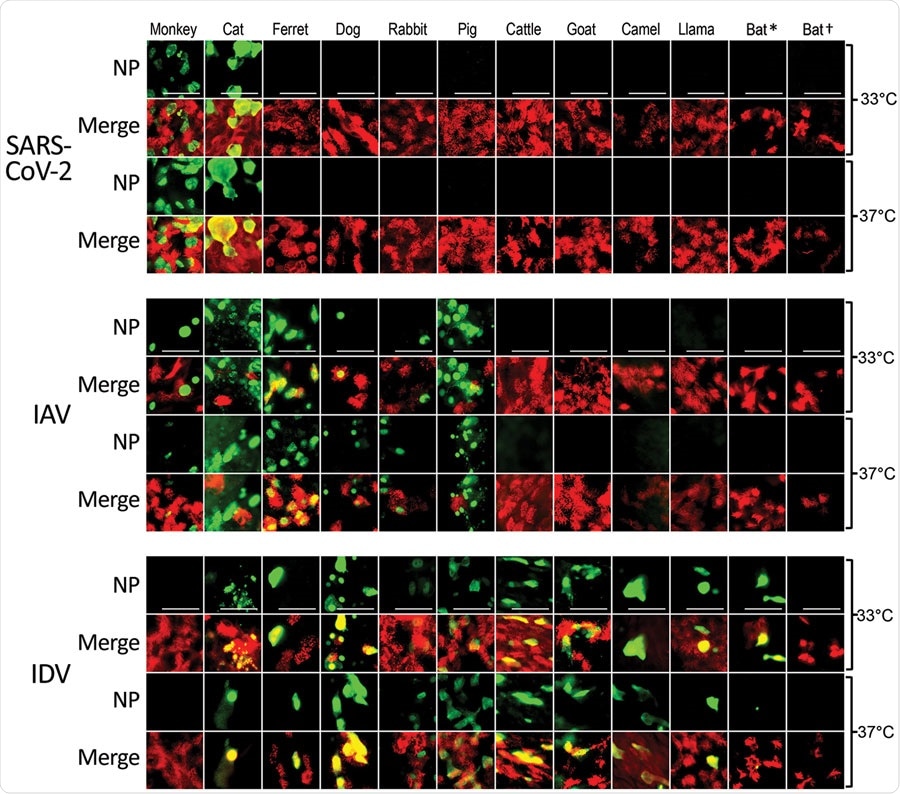
Tropisms of SARS-CoV-2, IAV, and IDV in infected airway epithelial cell cultures from diverse mammal species. We inoculated well-differentiated animal airway epithelial cell cultures with either 30.000 PFU of SARS-CoV-2 (SARS-CoV-2/München-1.1/2020/929), 10.000 50% tissue culture infective dose of IAV/Hamburg/4/2009, or IDV (D/bovine/Oklahoma/660/2013). We incubated virus-infected airway epithelial cell cultures at 33°C or 37°C and formalin-fixed them at 96 hours postinfection (for SARS-CoV-2) or 48 hours postinfection (for influenza viruses). After fixation, we stained virus-infected cultures using antibodies against either SARS-CoV-2, IAV, or IDV NP (green), and β-tubulin (cilia, red). We acquired images using an EVOS FL Auto 2 Imaging System equipped with a 40x air objective. Scale bar indicates 50 µm. *Sturnira lilium bat; †Carollia perspicillata bat. IAV, influenza A virus; IDV, influenza D virus; NP, nucleocapsid protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The team observed that SARS-CoV-2 replicated efficiently only in money and cat culture models. However, upon conducting whole-genome sequencing, they found no apparent signs of nucleotide transitions required for SARS-CoV-2 to infect monkey and cat airway epithelial cell (AEC) cultures productively.
The study findings suggest that certain species of these two animals may be particularly vulnerable to SARS-CoV-2 infection.
“Our findings, together with previous reports of human-to-animal spillover events, warrant close surveillance to determine the potential role of cats, monkeys, and closely related species as spillback reservoirs for SARS-CoV-2,” the team concluded in the study.
Further, the team says the study results highlight that in vitro well-differentiated animal AEC culture models can be used as an alternative to conventional animal experimentation models to study SARS-CoV-2’s host spectrum.
Journal reference:
- Gultom, M., Licheri, M., Laloli, L., Wider, M., Strassle, M. et al. (2021). Susceptibility of Well-Differentiated Airway Epithelial Cell Cultures from Domestic and Wild Animals to Severe Acute Respiratory Syndrome Coronavirus 2. Emerging Infectious Diseases. https://wwwnc.cdc.gov/eid/article/27/7/20-4660_article