Researchers in the United States have conducted a study showing that while variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can compromise the effectiveness of single therapeutic antibodies at preventing severe coronavirus disease 2019 (COVID-19), many combination antibody products remain potent against the variants.
Most clinical-stage therapeutic antibodies target the receptor-binding domain (RBD) of the viral spike protein that mediates the initial stage of the SARS-CoV-2 infection process when it binds to host cell receptors.
However, several viral variants have emerged containing multiple mutations in the spike that confer resistance to these antibodies.
To help predict this resistance, the researchers profiled the resistance patterns of 25 clinical-stage neutralizing antibodies (nAbs) against a large panel of variants containing single and multiple substitutions in the spike protein.
The team – from the US Food and Drug Administration (FDA) in Maryland, the US Department of Human Health and Services in Washington DC, and the National Institutes of Health in Maryland – found that 14 of 15 single antibodies were susceptible to at least one RBD substitution.
However, most combination nAb products and polyclonal nAbs retained their potency against the variants.
Carol Weiss and colleagues also found that the presence of critical substitutions in variants containing multiple spike substitutions were predictive of resistance.
However, the extent of this resistance could be modified in unpredictable ways by other spike substitutions lying outside of the RBD.
The researchers say the findings show that epistatic interactions in the spike protein can modify virus susceptibility to therapeutic antibodies, highlighting the importance of assessing antibody potency in the context of all substitutions present in a variant.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
More about antibody-based approaches to controlling the pandemic
Since the COVID-19 outbreak first began in late 2019, intense efforts to control SARS-CoV-2 transmission and prevent severe disease have led to the development of antibody-based countermeasures, including vaccines, therapeutic monoclonal antibodies and convalescent plasma.
Five nAb products that are in clinical use after having received emergency use authorization from the FDA target the SARS-CoV-2 spike protein.
Many of the current nAb products were developed using SARS-CoV-2 sequences that were isolated early on in the pandemic. However, the subsequent evolution of the virus has led to the emergence of distinct viral lineages containing key substitutions in the RBD that confer resistance to these nAbs.
One of the first variants to become dominant globally dominant contains the spike substitution D614G. Since then, the B.1.1.7 (alpha), B.1.351 (beta), P.1 (gamma) and A.23.1 lineages have also emerged in the United Kingdom, South Africa, Brazil, and Uganda, respectively.
Further variants of concern or variants of interest have also emerged regionally in the United States, including B.1.429/B.1.427 (epsilon) that was first identified in California and B.1.526 (iota) that was first identified in New York.
Ongoing assessments of nAbs are needed to ensure that they remain potent
“The continuing evolution of circulating SARS-CoV-2 variants, often containing substitutions known to confer resistance to several monoclonal antibodies, underscores the need for ongoing assessments of nAbs to ensure that they remain potent against new variants,” writes Weiss and colleagues.
“As it can take weeks to acquire new spike genes and viruses for performing in vitro evaluations, information about substitutions that can predict resistance to nAbs is needed,” they add.
What did the researchers do?
This team evaluated 25 nAbs, combination nAbs, or polyclonal nAbs currently in the late stages of clinical development for resistance against 60 pseudoviruses bearing spikes with single or multiple substitutions, including complete sets of mutations representing the B.1.1.7, B.1.351, P.1, A.23.1, B.1.429, and B.1.526 variants.
The team found that single substitutions in spike were sufficient to confer a loss of potency for 14 of 15 single nAbs.
Six of the 15 single nAbs lost potency against the RBD mutation E484K, which is present in multiple variant lineages.
However, most of the combination and polyclonal nAbs remained potent against the variants, emphasizing the importance of using combinations with complementary resistance profiles.
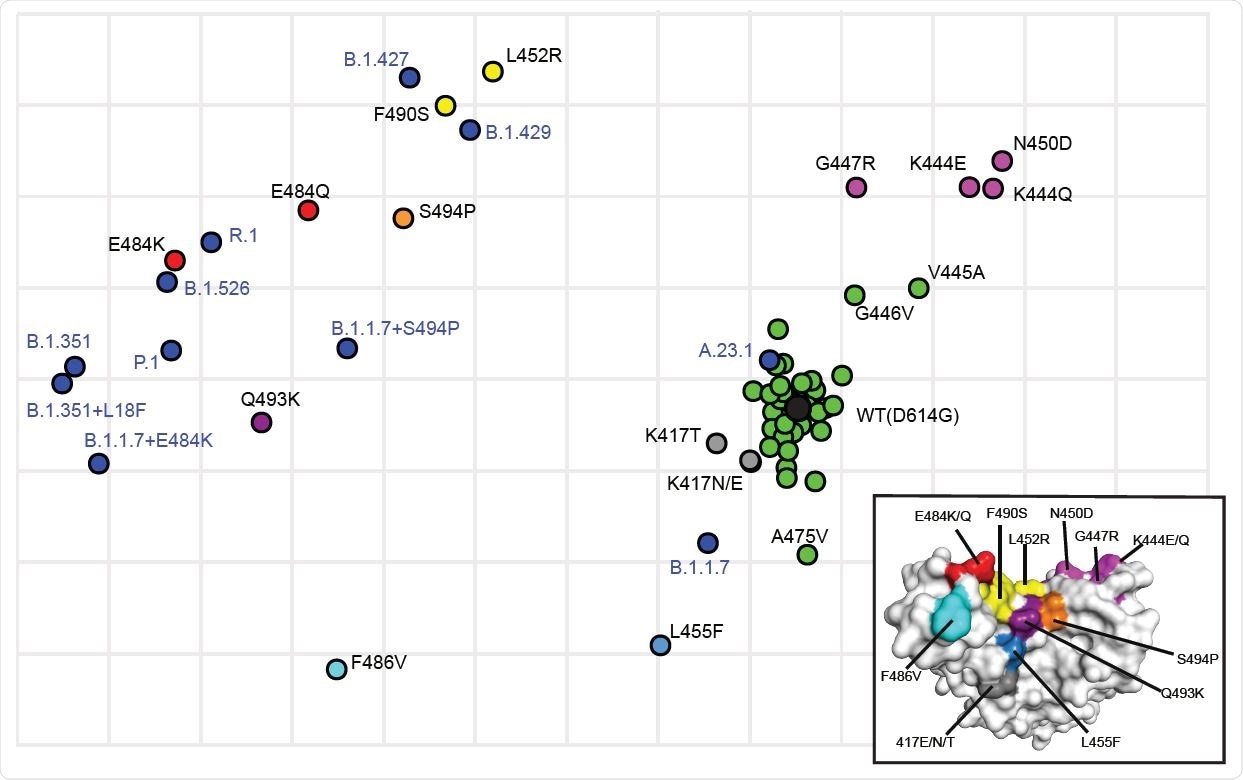
Antigenic cartography showing the relative antigenic distance of pseudoviruses with single substitutions in spike compared to spikes with the full set of substitutions found in variants. Antigenic maps were constructed using neutralization titers (dilution factors) of nAbs against all tested pseudoviruses. Blue dots identify pseudoviruses bearing Spikes representing variants of concern or interest. Black dot identifies the wild type (WT D614G) pseudovirus. Green dots identify pseudoviruses with single substitutions in Spike that are antigenically close to WT. Other colors identify pseudoviruses with single substitutions in Spike that are more antigenically distant from WT. Inset shows the color-coded locations of the single residue substitutions in the RBD.
Key substitutions predicted resistance to nAbs
The researchers also found that key substitutions in variants containing multiple spike mutations predicted resistance to the nAbs.
Furthermore, the team provided examples of the resistance conferred by a single RBD substitution being modified by other substitutions in the variant.
For one nAb, the resistance conferred by E484K was unexpectedly modified by the non-RBD substitution G769V or its combination with W152L, suggesting that long-range epistatic interactions occur in the spike.
“We note that a G769E substitution was previously identified in an escape mutant study involving selection with RBD antibodies,” writes Weiss and colleagues.
Antibody potency must be assessed in the context of all substitutions present in a variant
The researchers say these data highlight a role for epistatic interactions among residues within or outside the RBD that can affect antibody binding and potentially virus entry and evolution.
“These findings highlight the importance of assessing antibody potency in the context of all substitutions in a variant and show that epistatic interactions in spike can modify virus susceptibility to therapeutic antibodies,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Weiss C, et al. Key substitutions in the spike protein of SARS-CoV-2 variants can predict resistance to monoclonal antibodies, but other substitutions can modify the effects. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.07.16.452748, https://www.biorxiv.org/content/10.1101/2021.07.16.452748v1
- Peer reviewed and published scientific report.
Lusvarghi, Sabrina, Wei Wang, Rachel Herrup, Sabari Nath Neerukonda, Russell Vassell, Lisa Bentley, Ann E. Eakin, Karl J. Erlandson, and Carol D. Weiss. 2022. “Key Substitutions in the Spike Protein of SARS-CoV-2 Variants Can Predict Resistance to Monoclonal Antibodies, but Other Substitutions Can Modify the Effects.” Edited by Kanta Subbarao. Journal of Virology 96 (1). https://doi.org/10.1128/jvi.01110-21. https://journals.asm.org/doi/10.1128/JVI.01110-21.