The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), continues to challenge normal social and economic activity.
The most baffling aspect of COVID-19 is its apparent unpredictability. While causing no or mild symptoms in a majority of infected individuals, COVID-19 can alternatively cause dysregulated immune inflammation, which can lead to systemic damage and even death in many others.
 Study: HLA-dependent variation in SARS-CoV-2 CD8+ T cell cross-reactivity with human coronaviruses. Image Credit: Design_Cells / Shutterstock.com
Study: HLA-dependent variation in SARS-CoV-2 CD8+ T cell cross-reactivity with human coronaviruses. Image Credit: Design_Cells / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A new study published on the preprint server bioRxiv* discusses how both different responses to the infection itself and to COVID-19 vaccines are likely the result of genetic differences. These differences appear to impact the nature of pre-existing immunity to SARS-CoV-2 that had been induced by prior infections with other endemic seasonal human coronaviruses (hCoVs).
Background
Cross-reactivity between SARS-CoV-2 antigens and those of other hCoVs could enhance the immune response to natural infection or vaccination, or reduce the severity of illness, through memory T cells. However, this cross-reactivity can also cause antibody-dependent enhancement (ADE) of infection or promote the generation of antibodies, which can lead to chronic illness.
Four endemic hCoVs are known, of which include HCoV-229E, -NL63, -OC43, and -HKU1. Earlier studies have shown that SARS-CoV-2-reactive T cells, particularly CD4+ and CD8_ cells, have been found in uninfected individuals.
Evidence of CD8+ T cell immunity to the virus in naïve individuals is more controversial. The current study indicates that there is a strong probability that memory T cells elicited by previous hCoV infections can react against highly similar SARS-CoV-2 antigens, conferring protection against the latter virus. However, this is dependent on the presence of specific cell recognition proteins known as the human leukocyte antigen (HLA) cells.
What did the study show?
The researchers found thousands of immunogenic SARS-CoV-2 peptide antigens that were presented by class I and class II major histocompatibility complexes (MHC). Both class I and class II MHC molecules comprised about a third of these antigens each, while MHC type for the remaining third was not known. The HLA alleles for the MHC complexes were known in approximately 57% and 42%, respectively.
By focusing on the immunogenic peptides, the researchers successfully identified the missing information. Through the analysis of HLA preferences towards the viral peptides, the researchers found that among 9-mers, HLA-C*07:02 was seen most frequently.
A total of 126 SARS-CoV-2 peptides presented with high similarity to almost 300 HCoV peptides. The former were presented by multiple HLA alleles, over half of which were 9-mers, a fifth of them 15-mers, and over one in seven 10-mers.
The greatest number of matches was with HKU1 and OC43, which is likely due to their shared total similarity with the SARS-CoV-2 genome. In addition, 42 SARS-CoV-2 peptides that bound to three or more hCoVs, along with small groups, showed homology to only one strain.
Shared peptides were most abundant with Viral ORF1ab and the spike proteins for OC43 and HKU1. These findings related to sequences with 70% or more homology. The findings suggest that there are potential approaches to protecting against this virus via pre-existing immunity.
Dissimilarity to the human proteome
The researchers found that SARS-CoV-2 peptides presented by class I HLA had greater dissimilarity to the human proteome when they were immunogenic vs non-immunogenic. This was not the case with class II-presented peptides.
To this end, the authors of the current study detected several immunogenic SARS-CoV-2 peptides with a mean similarity of 72% to self-antigens. The T cells that are reactive to these peptides should therefore be negatively selected to prevent autoimmune pathology or ADE. Among these includes CCL3 and CCL3L1, both of which are commonly elevated during the COVID-19 cytokine storm.
The peptides that showed homology to these signaling molecules were part of the private T cell repertoire, potentially enhancing the risk of immunopathology. Over 70% and almost 80% of amino acids were conserved between 10- and 9-mers, respectively.
These peptides deserve to be further investigated for their role in potential autoimmunity after vaccination or natural infection with COVID-19.
CD8+ T cell cross-reactivity
Nearly 250 SARS-CoV-2 peptides have specific T cell receptors. Interestingly, 80% of TCRs recognize 16% (n=40) viral peptides, 36 being private and the rest shared by both clusters. Notably, the V gene TRBV20-1 and the J gene TRBJ2-1 were found to be over-represented.
Graphing their interactions showed two clear hubs, with healthy people being connected to public TCRs recognizing shared peptides or SARS-CoV-2 peptides (mostly the former) on one hub. The second hub, representing convalescent COVID-19 patients, showed mostly public TCR repertoires with their cognate SARS-CoV-2-private peptides.
Many TCRs recognized peptides common to both these groups, showing the presence of public TCRs to shared peptides in naïve individuals as well as COVID-19 patients. This may not indicate the presence of pre-existing T cell immunity, but rather of cross-reactivity.
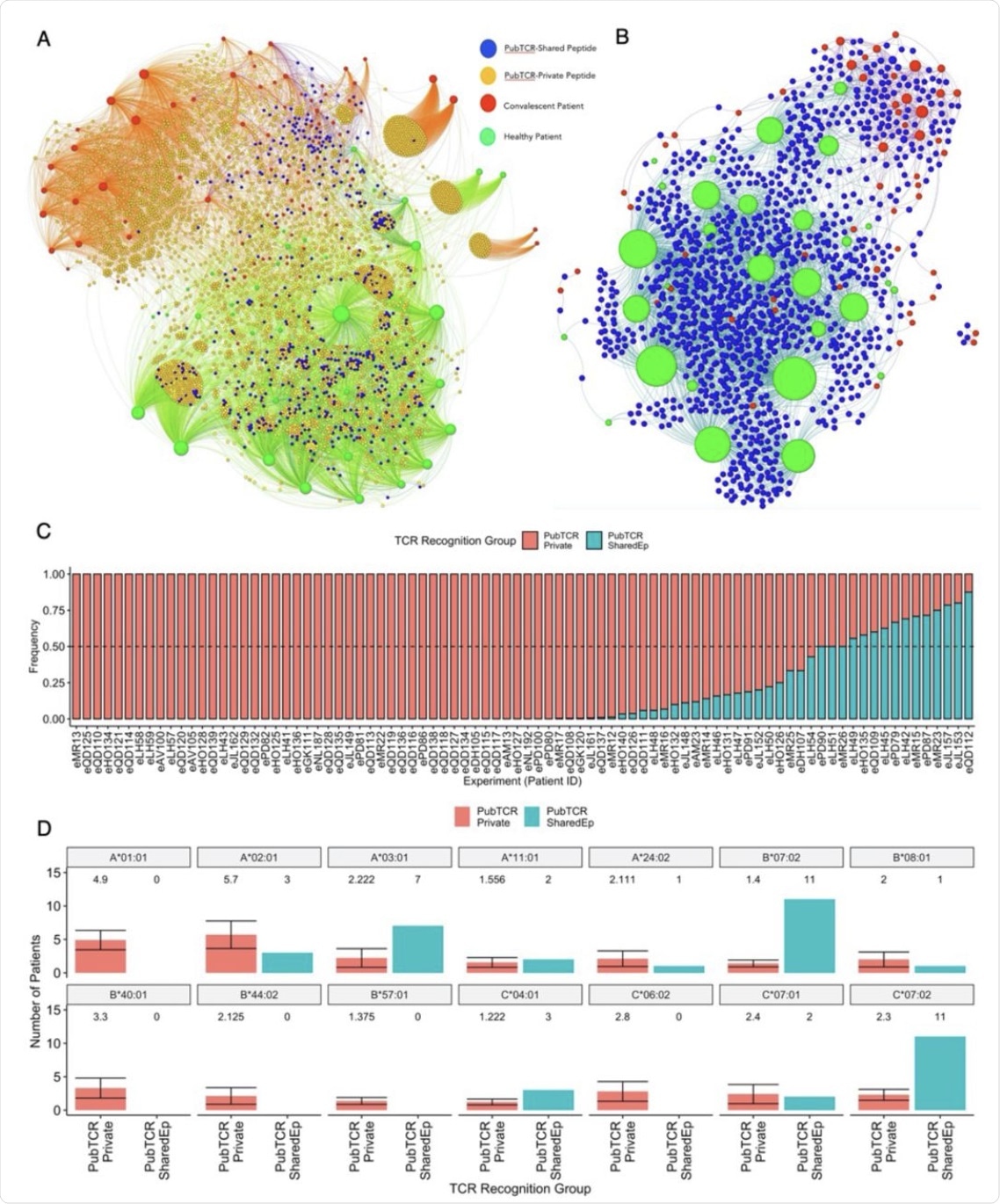 A) A bipartite network graph showing SARS-CoV-2-specific public TCRs which recognize shared or private SARS-CoV-2 peptides in healthy or convalescent patients. TCRs are color-coded by whether they recognize only shared peptides (blue), only private peptides (orange) or both (yellow). COVID-19 convalescent patients are labeled red while healthy controls are labeled green. Node size reflects the degree of connectivity, i.e. the quantity of an individual’s TCRs that are shared with other patients. B) A bipartite network graph showing SARS-CoV-2 public TCRs that recognize SARS-CoV-2-HCoV shared peptides. Patient node size reflects the quantity of their TCRs which are shared with another patient. Healthy patients are labeled green, COVID-19 convalescent is labeled red, and (public) TCRs are labeled blue. C) A bar plot showing the frequency that each convalescent patient’s public TCRs recognize SARS-CoV-2-private (red) or SARS-CoV-2-HCoV-shared (blue) peptides. Patients with identical frequencies are ordered by the number of TCRs. D) Barplots showing the quantities of COVID-19 convalescent patients who carry 14 class I HLA alleles of interest. Patients are grouped by whether their public TCRs predominately recognize “Private” (PubTCR_Private, n=12, sampled 10 times) or “Shared” (PubTCR_SharedEp, n=12) peptides. For the PubTCR-Private group, 12 patients were sampled 10 times and the number of patients carrying alleles was measured. The mean number of patients carrying each allele and the error are visualized. For the PubTCR-SharedEp group, the data contain only 12 patients of interest, thus the number of patients carrying each allele is measured and visualised.
A) A bipartite network graph showing SARS-CoV-2-specific public TCRs which recognize shared or private SARS-CoV-2 peptides in healthy or convalescent patients. TCRs are color-coded by whether they recognize only shared peptides (blue), only private peptides (orange) or both (yellow). COVID-19 convalescent patients are labeled red while healthy controls are labeled green. Node size reflects the degree of connectivity, i.e. the quantity of an individual’s TCRs that are shared with other patients. B) A bipartite network graph showing SARS-CoV-2 public TCRs that recognize SARS-CoV-2-HCoV shared peptides. Patient node size reflects the quantity of their TCRs which are shared with another patient. Healthy patients are labeled green, COVID-19 convalescent is labeled red, and (public) TCRs are labeled blue. C) A bar plot showing the frequency that each convalescent patient’s public TCRs recognize SARS-CoV-2-private (red) or SARS-CoV-2-HCoV-shared (blue) peptides. Patients with identical frequencies are ordered by the number of TCRs. D) Barplots showing the quantities of COVID-19 convalescent patients who carry 14 class I HLA alleles of interest. Patients are grouped by whether their public TCRs predominately recognize “Private” (PubTCR_Private, n=12, sampled 10 times) or “Shared” (PubTCR_SharedEp, n=12) peptides. For the PubTCR-Private group, 12 patients were sampled 10 times and the number of patients carrying alleles was measured. The mean number of patients carrying each allele and the error are visualized. For the PubTCR-SharedEp group, the data contain only 12 patients of interest, thus the number of patients carrying each allele is measured and visualised.
It is also observed that while most (51/86) convalescent patients have public TCRs binding to primarily private SARS-CoV-2 peptides, 12 patients exhibited more than 50% of these TCRs that are capable of recognizing shared peptides. These patients showed HLA-B*07:02, HLA-C*07:02, and HLA A*03:01 enrichment, which was comparable to a broader HLA allele set, with a higher representation of HLA A*01:01, in the private peptide group.
Taken together, these findings corroborate previous reports demonstrating that CD8+ T Cell HCoV-CoV-2 cross-reactivity may be conditioned by HLA. Thus, only these patients may have stronger protection against SARS-CoV-2.
These findings show that there is a set of epitopes with cognate public TCRs in healthy controls, along with a novel set of peptides that evoke T cell responses in COVID-19. The latter peptide set is not seen to be expanded in unexposed patients.
However, there are also private TCRs in naïve subjects with cognate homologous peptides of both SARS-CoV-2 and the hCoVs. Some of these are actually public TCRs, having been reported in independent studies, and capable of cross-reacting with both SARS-CoV-2 and OC43/HKU1 homologous peptides.
Identifying conserved peptides for protective immunity
The scientists also found that a total of 84 peptides are cognate to TCRs in convalescent and naïve individuals. They then identified ten SARS-CoV-2 peptides that were very similar to the earlier SARS-like CoVs. These peptides will likely be presented by multiple HLA alleles and may therefore be responsible for a T cell response among 93% of the world’s population.
What are the implications?
“We believe that this is an encouraging insight in the search for pan-coronavirus T cell targets, and additionally propose these as top candidates for cross-protective immunity.”
Future work will be required to fully understand how immunity is improved by cross-reactive T cell responses. It is also necessary to explore if cross-reactive TCRs will be induced by COVID-19 vaccines, as this may assist clinicians in identifying which patients will develop cross-reactive responses after vaccination.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Buckley, P. R., Lee, C. H., Pinho, M. P., et al. (2021). HLA-dependent variation in SARS-CoV-2 CD8+ T cell cross-reactivity with human coronaviruses. bioRxiv. doi:10.1101/2021.07.17.452778. https://www.biorxiv.org/content/10.1101/2021.07.17.452778v1.full
- Peer reviewed and published scientific report.
Buckley, Paul R., Chloe H. Lee, Mariana Pereira Pinho, Rosana Ottakandathil Babu, Jeongmin Woo, Agne Antanaviciute, Alison Simmons, Graham Ogg, and Hashem Koohy. 2022. “HLA‐Dependent Variation in SARS‐CoV‐2 CD8 + T Cell Cross‐Reactivity with Human Coronaviruses.” Immunology 166 (1): 78–103. https://doi.org/10.1111/imm.13451. https://onlinelibrary.wiley.com/doi/10.1111/imm.13451.