A new genetic study from an international team of scientists has uncovered genes involved in severe coronavirus disease 2019 (COVID-19) illness. Two loci at 17q21.31 and 19q13.33 are associated with respiratory failure during infection.
The loci are thought to cause impairments in lung function and alter the number of immune and blood cells. The genetic changes also affect the NAPSA gene, which contributes to lung surfactant protein production.
Led by Andre Franke from Christian-Albrechts-University and University Hospital Schleswig-Holstein in Germany, the results provide insight into how genetics contribute to COVID-19 disease severity. According to the authors, their findings could aid with designing more targeted experiments focusing on specific genes and subsequently develop therapies to alleviate severe COVID-19 infection.
The study “New susceptibility loci for severe COVID-19 by detailed GWAS analysis in European populations” is published and available on the preprint medRxiv* server.
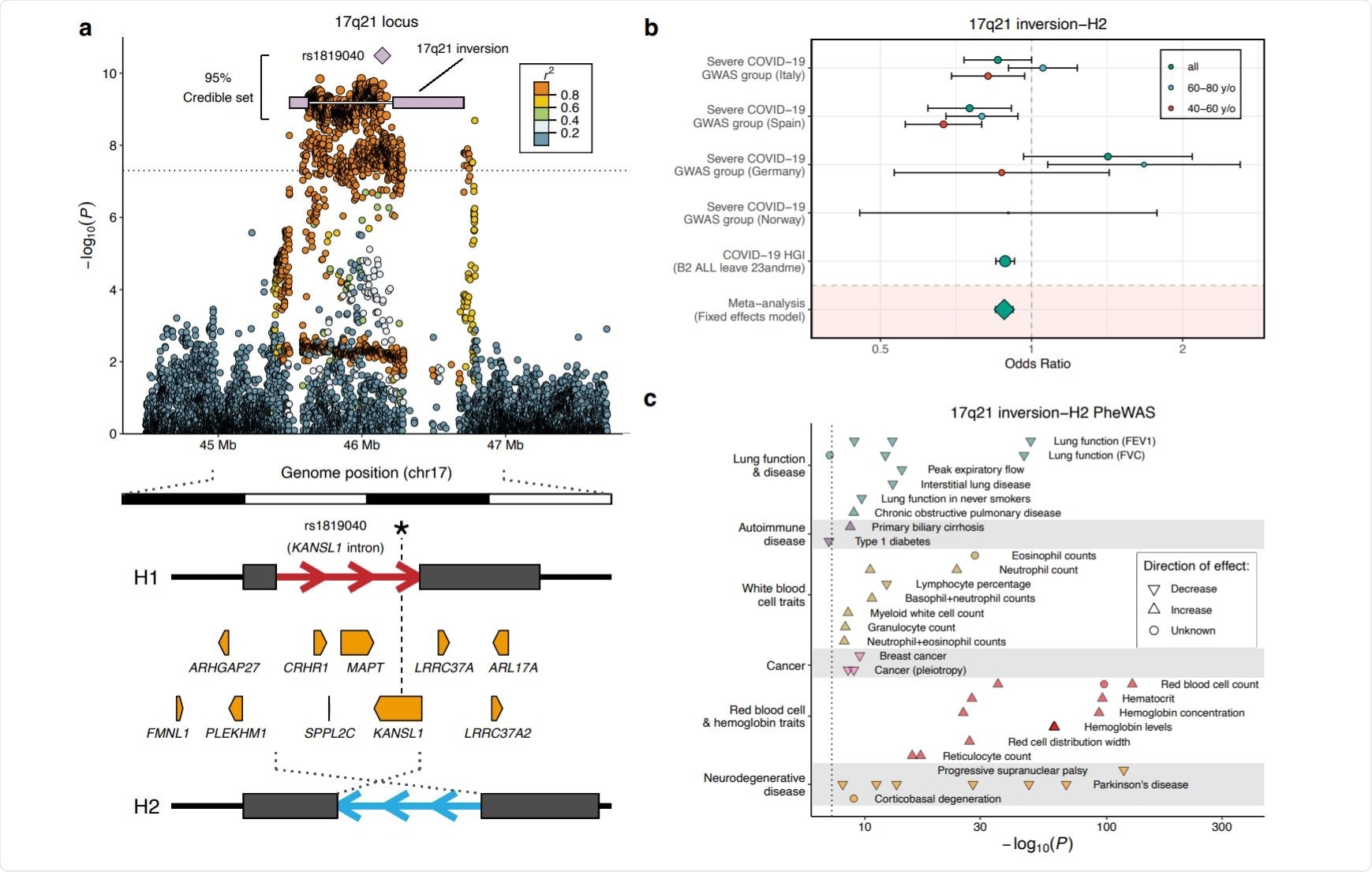
Image Credit: New susceptibility loci for severe COVID-19 by detailed GWAS analysis in European populations

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The course of the experiment
The researchers conducted a meta-analysis looking at genetic loci from patients with severe COVID-19 illness living in Italy, Spain, Norway, and Germany/Austria. Their focus was on genetic variations on the human leukocyte antigens (HLA) and how they may play a role in disease severity.
Another point of interest for the researchers was to look for a possible genetic link between severe COVID-19 and the male sex. Research suggests men are more likely to develop more severe infections than women. To this end, the team investigated genetic variations on the Y chromosome with a focus on the haplogroup R — the predominant haplogroup in Europe.
Genes contribute to COVID-19 disease severity
The genetic analysis identified 17q21.31 inversion and 19q13.33 as two loci linked to severe COVID-19 infection.
The 19q13.33 locus appeared to contribute to severe disease infection by affecting its regulation of the NAPSA gene. The gene helps with the production of an enzyme expressed in Type 1 and Type 2 alveolar cells. These cells are needed for exchanging gases at the lung surface, secreting surfactant proteins, and immunomodulatory factors.
The NAPSA gene was associated with damage-associated transient progenitors, an intermediate state between cell types with high inflammation during COVID-19 infection.
“Thus, given that the NAPSA protein is involved in lung surfactant production, which is dysregulated in COVID-19, the fact that the COVID-19 risk allele is associated with decreased NAPSA expression while increased expression of NAPSA is a protective factor for severe COVID-19, suggests a potential role of NAPSA in susceptibility for severe COVID-19,”
Role of genetic inversions in COVID-19 illness
The 17q21.31 inversion may dysregulate the immune response during COVID-19 infection.
Previous studies have suggested the inversion is linked to a higher red blood cell count and elevated hemoglobin levels. Additionally, it correlates with a decreased lung function and a higher risk of developing the chronic obstructive pulmonary disease.
This genetic locus was of interest because it is commonly expressed among Europeans.
Although the function of many of the affected genes are not well known, the inversion acts as an eQTL and sQTL of several interesting candidate genes for severe COVID-19.
Other genes that could impair immune function and ultimately result in more severe infection include KANSL1, CHRH1, and FMNL1. The KANSL1 gene contributed to histone acetylation and was found to exhibit decreased expression in infection-like stimulated monocytes, which is “partially compensated in homozygotes for the H2 inversion haplotype.”
The CHRH1 gene showed greater expression in the H2 haplotype in multiple tissues. The researchers suggest it could regulate inflammatory responses through its involvement with corticotropin-releasing hormones.
The FMNL1 gene is also found on the inversion locus and is highly expressed in macrophages, dendritic cells, B lymphocytes, and T lymphocytes in tissues infected with COVID-19.
Further genetic analysis also found a downregulation of the MAPT genes in neurons and lung cells infected with the coronavirus. Though the 17q21.31 inversion is associated with increased expression of MAPT in several tissues, suggesting a protective effect against SARS-CoV-2. But the researchers clarify that more research is needed as it is unlikely to pick out which gene is the most protective or detrimental during COVID-19 infection.
No significant genetic link in Y-chromosome haplogroups
The study did not yield any significant association between severe COVID-19 infection and genetic variations on the Y chromosome.
Although the researchers note that larger samples may have been needed to study Y-chromosome haplogroups and their connection to COVID-19 disease.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Source:
Journal references:
- Preliminary scientific report.
Degenhardt F, et al. New susceptibility loci for severe COVID-19 by detailed GWAS analysis in European populations. medRxiv, 2021. doi: https://doi.org/10.1101/2021.07.21.21260624, https://www.medrxiv.org/content/10.1101/2021.07.21.21260624v1
- Peer reviewed and published scientific report.
Degenhardt, Frauke, David Ellinghaus, Simonas Juzenas, Jon Lerga-Jaso, Mareike Wendorff, Douglas Maya-Miles, Florian Uellendahl-Werth, et al. 2022. “Detailed Stratified GWAS Analysis for Severe COVID-19 in Four European Populations.” Human Molecular Genetics, July, ddac158. https://doi.org/10.1093/hmg/ddac158. https://academic.oup.com/hmg/article/31/23/3945/6644888.