The global outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in the coronavirus disease 2019 (COVID-19) pandemic. To date, this pandemic has claimed more than 4 million lives worldwide and infected over 194 million. Scientists around the world are conducting extensive research to understand the characteristic features of this virus and its modes of infection.
One of the most important structural proteins of SARS-CoV-2 is the spike (S) protein. To this end, the S protein has gained considerable attention owing to its role in the entry of SARS-CoV-2 into host cells and tissue tropism. As a result, the S protein is the main antigenic target of all COVID-19 vaccines authorized for emergency use.
Recent research using cryogenic electron microscopy (cryo-EM) has revealed that the S protein is present on the virion as a homotrimer of approximately 180 kDa per protomer. Each protomer consists of two subunits known as S1 and S2.
S1 contains the receptor-binding domain (RBD) that binds with the host’s angiotensin-converting enzyme 2 (ACE2) receptor. Comparably, S2 is responsible for the fusion of the virus envelope with the host cell membrane.
During the invasion of SARS-CoV-2 into the host cell, the S protein undergoes glycosylation with 22 N-glycosylation sites per protomer. A glycan shield is formed, owing to the heavy glycosylation, which helps the virus to evade antigenic recognition. The glycan shield also stabilizes the RBD to promote favorable binding with the ACE2 receptor.
The mechanism behind the increased virulence of the VoCs
Postemergence of SARS-CoV-2 variants, researchers have sequenced the entire genome of the virus to study the genomic changes. Researchers have reported amino acid substitutions in the S protein region.
Some of the SARS-CoV-2 variants including the Alpha, Gamma, Beta, and Delta variants, have been referred to as Variants of Concern (VoC). Others, such as Epsilon, Iota, and Kappa variants, have remained Variants of Interest (VoI).
Earlier studies associated with S mutations have shown that S-D614G mutated variants have increased infectivity in human lung cells coupled as demonstrated by an enhanced rate of viral replication. Cryo-EM data, along with molecular dynamic (MD) studies, have revealed that the SD614G mutation prefers “open” conformations as compared to the wild type.
SARS-CoV-2 variants can disrupt the interchain associations made via hydrogen bonds between D614 and residue T859 on an adjacent molecule. These changes bring the RBD in an “up” position, which promotes its interaction with the ACE2 receptor. MD studies have also indicated that glycans on N165 and N234 stabilize the open form, thereby increasing RBD interactions with ACE2.
Mass spectrometry to analyze protein glycosylation
In both global proteomics analyses and targeted studies related to vaccinology; mass spectrometry (MS) is the preferred technique for its site-specific analysis of the protein glycosylation. To this end, MS can determine the glycan composition and its molecular size, the fragmentation pattern of a glycopeptide, and the precise site of glycosylation.
A new study published on the preprint server bioRxiv* has characterized and compared the N-glycosylation of the wild type (S-614D) and variant (S-614G) of SARS-CoV-2 S glycoproteins using MS techniques.
In their work, the authors used a glycoproteomics approach based on signature ions triggered electron-transfer dissociation MS to characterize the S proteins of SARS-CoV-1 and -2, as well as the Middle East Respiratory Syndrome (MERS) from multiple expression systems. The glycan structures of these S proteins were found to be dependent on the expression system used to produce the protein and the structure of the construct.
Study findings
In the current study, the researchers presented the N-glycosylation profiles of the SARS-CoV-2 spike D614G variant and its ancestor protein using advanced EThcD MS.
To obtain accurate results, the researchers constructed recombinant proteins using identical cells that were expressed under identical conditions. About 50% of the N-glycosylation sequons were found to have altered their distribution of glycans in the S-614G variant. Further, a decrease in the relative abundance of complex-type glycans and an increase in oligomannose glycans on all changed sequons of the S-614G variant was observed.
These findings indicate a reduction in the overall complexity of the total N-glycosylation profile on the S-614G protein, which plays a key role in reshaping the homotrimer. Modification in the N-glycan occurs on the head and stalk of the ectodomain S protein. Interestingly, this study revealed that all the altered patterns in the glycosylation sites were found in the S head, whereas the glycosylation of three sites in the stalk portion remained unchanged between S-614G and S-614D proteins.
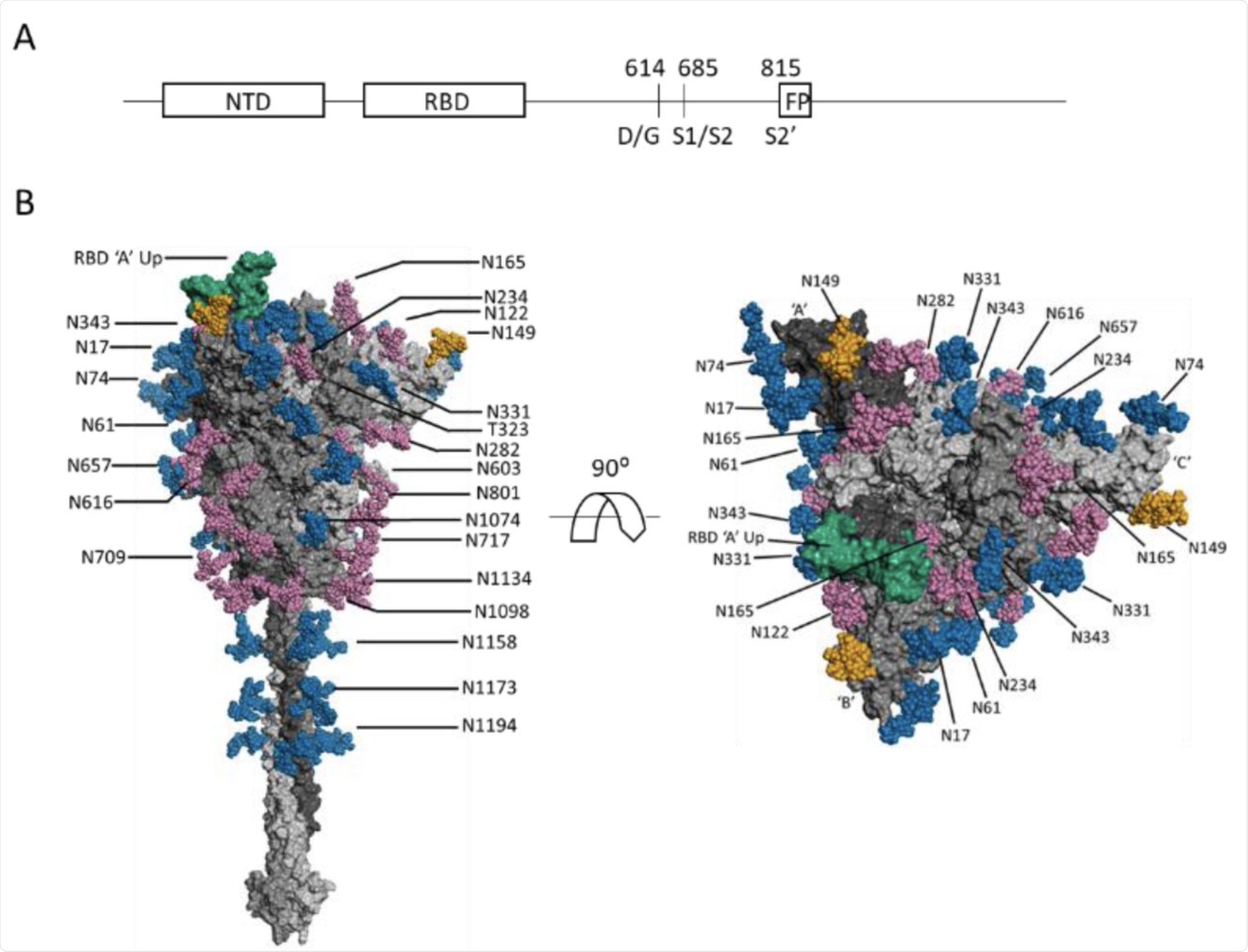 (A) Schematic SARS-CoV-2 S protein primary structure. (B) N-glycans depicted on a representative full-length, fully-glycosylated prefusion conformation of the trimetric SARS-CoV-2 spike protein (file 6vsb 1 1 1.pdb from the CHARMM-GUI Archive displayed in PyMOL). Blue-colored glycans indicate no change in the glycosylation site between the S-614G mutant and the S-614D wild type. Magenta-colored glycans indicate a modification in the glycan distribution and type between the mutant and wild type. The RBD is shown in green. The N-149 glycans are gold. The glycans depicted do not necessarily match those described in this report, and the O-linked glycans in the model are hidden due to low occupancy.
(A) Schematic SARS-CoV-2 S protein primary structure. (B) N-glycans depicted on a representative full-length, fully-glycosylated prefusion conformation of the trimetric SARS-CoV-2 spike protein (file 6vsb 1 1 1.pdb from the CHARMM-GUI Archive displayed in PyMOL). Blue-colored glycans indicate no change in the glycosylation site between the S-614G mutant and the S-614D wild type. Magenta-colored glycans indicate a modification in the glycan distribution and type between the mutant and wild type. The RBD is shown in green. The N-149 glycans are gold. The glycans depicted do not necessarily match those described in this report, and the O-linked glycans in the model are hidden due to low occupancy.
Conclusion
The current study has provided evidence to assist in understanding the effects of protein glycosylation on the structure and function of the S protein of SARS-CoV-2.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Wang, D., Zhou, B., Keppel, T., et al. (2021). N-glycosylation profiles of the SARS-CoV-2 spike D614G mutant and its ancestral protein characterized by advanced mass spectrometry. bioRxiv. doi:10.1101/2021.07.26.453787. https://www.biorxiv.org/content/10.1101/2021.07.26.453787v1
- Peer reviewed and published scientific report.
Wang, Dongxia, Bin Zhou, Theodore R. Keppel, Maria Solano, Jakub Baudys, Jason Goldstein, M. G. Finn, et al. 2021. “N-Glycosylation Profiles of the SARS-CoV-2 Spike D614G Mutant and Its Ancestral Protein Characterized by Advanced Mass Spectrometry.” Scientific Reports 11 (1): 23561. https://doi.org/10.1038/s41598-021-02904-w. https://www.nature.com/articles/s41598-021-02904-w.