Since the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Wuhan, China at the end of 2019, several new variants have also been identified. As of July 29, 2021, the coronavirus disease 2019 (COVID-19), which is the disease caused by SARS-CoV-2, has caused almost 4.2 million deaths and infected over 195 million worldwide.
The lack of safe and effective antivirals to treat COVID-19 has led to an intensive global effort to develop an effective vaccine. Despite the rollout of several vaccines that elicit a robust immune response against the SARS-CoV-2 wildtype (WT) strain, the persistent emergence of new variants poses a formidable challenge to immune neutralization of the virus by vaccination.
 Study: Differential Interactions Between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. Image Credit: Fit Ztudio / Shutterstock.com
Study: Differential Interactions Between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. Image Credit: Fit Ztudio / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
SARS-CoV-2 gains entry to the host cell via its spike protein. Through the spike protein’s receptor-binding domain (RBD), this protein interacts with the host cell angiotensin-converting enzyme 2 (ACE2) receptor.
The ACE2 receptor was also utilized by SARS-CoV-1 to enter the host cell, which was circulating around the world in 2002. Therefore, the interactions between this receptor and these two coronaviruses could help understand the nature of binding as well as the differences between the agents themselves.
The Alpha (B.1.1.7 lineage) variant of SARS-CoV-2, which was first detected in England, spread so rapidly that it quickly became the dominant circulating strain of SARS-CoV-2. It has since been detected in over a hundred countries worldwide. The Beta variant (B.1.351 lineage), which was first identified in South Africa, also quickly spread to many nations around the world, including the United States.
Other SARS-CoV-2 variants that have been identified since the start of the COVID-19 pandemic include the Gamma (P.1), Epsilon (B.1.427), Kappa (B.1.617.1), and Delta (lineage B.1.617.2) variants, the latter of which is currently the dominant strain circulating in the United States. Taken together, all of these SARS-CoV-2 variants have been described as variants of concern (VoCs) by the United States Centers for Disease Control and Prevention (CDC).
Earlier studies have examined infectivity characteristics using both experimental and computational methods, which suggest that RBD-ACE2 binding is mediated by π-π interactions and π-cation interactions. Important residues involved in this binding include F486, Q498, T500, and Y505, all of which are located in the RBD.
How was the study done?
The current study, using all-atom steered molecular dynamics (SMD) simulations and microscale thermophoresis (MST) experiments, explored ACE2-RBD interactions with four VoCs including the Alpha, Beta, Gamma, and Delta variants, as well as the Epsilon and Kappa, both of which are variants of interest (VoIs).
The researchers modeled the pulling force on the fully glycosylated spike RBD-ACE2 model. Most variants showed high force profiles compared to the WT virus, except for the epsilon variant., thus indicating that these variants bind more strongly to the receptor than the parental strain.
Greatest pulling force for the Alpha variant
The highest pulling force that successfully drew the RBD-ACE2 complex in the opposite direction was required for the Alpha variant. This was likely because the RBD Y501 interacts more strongly with the ACE2 Q42, Y41, and D38. This is unlike the bonds between the WT or epsilon RBD variants, which do not have the N501Y mutation.
The researchers also found that the RBD N501Y mutation resulted in 40% more contacts occurring within 4.5 Å of the heavy atoms of the ACE2 residues that mediated its interactions with the RBD.
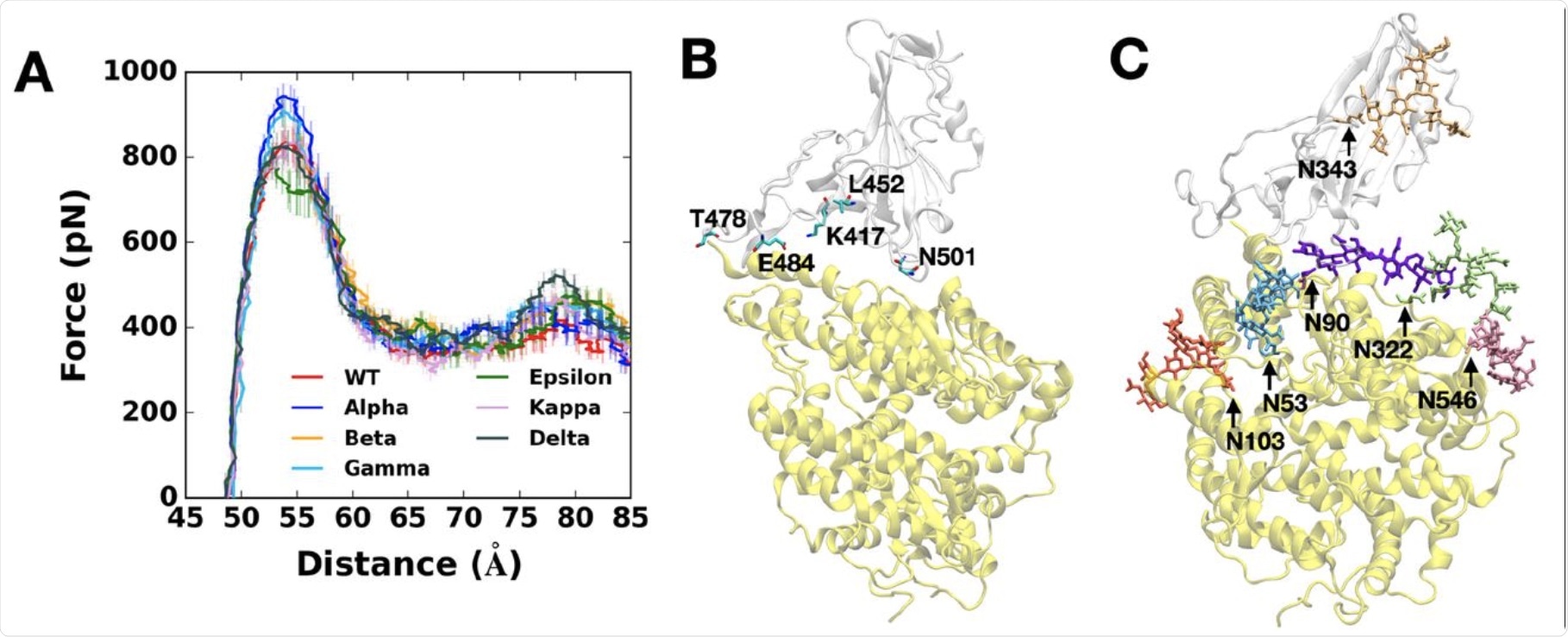 (A) Average force profiles of WT (red), Alpha (blue), Beta (orange), Gamma (sky blue), Epsilon (green), Kappa (pink), and Delta (gray) variants as a function of the distance between the centers of mass of RBD and ACE2. (B) Initial snapshot of WT. Residues subjected to each mutation are shown as solid sticks (N501, K417, E484, L452, and T478). RBD and ACE2 are respectively colored in light gray and yellow. All N-glycans, water, and ions are hidden for clarity. (C) Initial snapshot of WT with clockwise 90° rotation along the normal from (B). All N-glycans are depicted in different colors. Any other residues, water, and ions are not shown for clarity.
(A) Average force profiles of WT (red), Alpha (blue), Beta (orange), Gamma (sky blue), Epsilon (green), Kappa (pink), and Delta (gray) variants as a function of the distance between the centers of mass of RBD and ACE2. (B) Initial snapshot of WT. Residues subjected to each mutation are shown as solid sticks (N501, K417, E484, L452, and T478). RBD and ACE2 are respectively colored in light gray and yellow. All N-glycans, water, and ions are hidden for clarity. (C) Initial snapshot of WT with clockwise 90° rotation along the normal from (B). All N-glycans are depicted in different colors. Any other residues, water, and ions are not shown for clarity.
In particular, the Alpha variant’s Y501 was closer to the Y41 and K353 residues on the ACE2 protein. Again, the RBD of the Alpha variant has the most contact with the N90 glycan residue on the ACE2 and, as a result, its rapid rise to dominance.
The pulling force required for the Beta and Gamma variants was weaker as compared to that of the Alpha variant, though still higher than that which was required with the WT variant.
Less transmissible VoCs
The Beta variant also contains the N501Y mutation, in addition to the K417N and E484K mutations. The Y501 mutation interacts at a higher level with the D38, Y41, and Q42 on the ACE2 receptor, but with a lower frequency between the RBD N417 and ACE2 D30/H34, as well as RBD K484 and ACE2 K31.
Similar to the Beta variant, the Gamma variant also has lower contacts due to the K417T mutation, which is the sole difference between the two (vs K417N in the Beta lineage). However, it was observed that, unlike the Beta variant, all other variants with the K417 residue had RBD-ACE2 contacts between 50 to 60 Å.
This could be because the shortened side chain, as a result of the substitution of lysine to asparagine, may impair RBD-ACE2 interactions by causing alterations at their interface. With the Gamma variant, which has an even shorter threonine residue at this residue (T417), there is almost no interaction.
Thus, the RBD of both the Beta and Gamma variants, which contain N417 and T417, respectively, may be less contagious than the Alpha variant. Conversely, the N501Y mutation allowed both of these variants to interact with the ACE2, perhaps accounting for the rise in prevalence of the Gamma variant to the second-highest place among the different VoCs and VoIs.
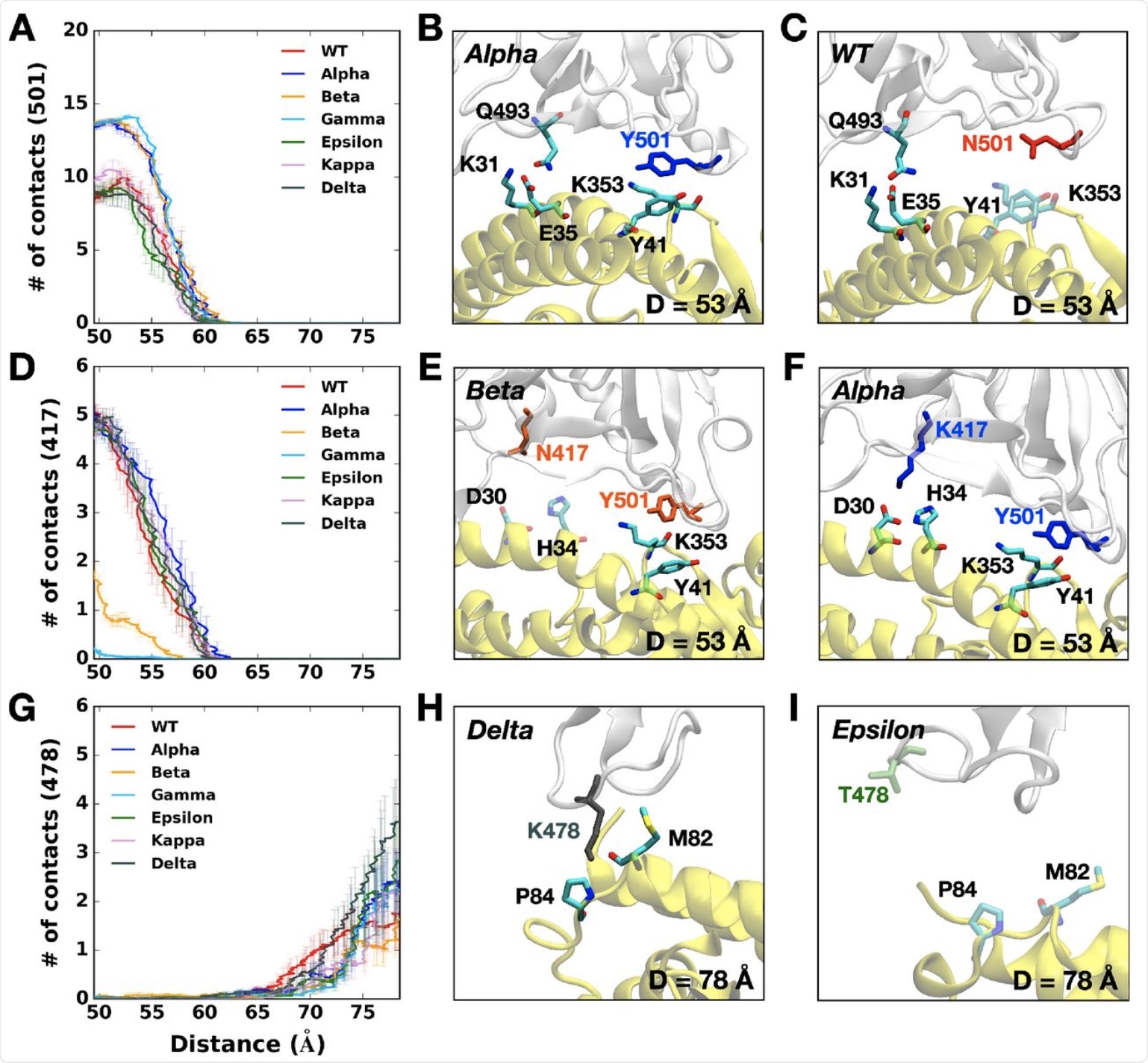 (A) The average number of contacts between RBD residue 501 and ACE2. (B and C) Representative snapshots at D = 53 Å of (B) Alpha variant and (C) WT. (D) The average number of contacts between RBD residue 417 and ACE2 and (E and F) their interacting residue pairs at D = 53 Å of (E) Beta and (F) Alpha variants. (G) The average number of contacts between RBD residue 478 and ACE2 and (H and I) key interaction pairs at D = 78 Å of (H) Delta and (I) Epsilon variants. The overall color scheme is the same as in Figure 1, and each mutated residue in each variant is shown using the same colors (i.e., red for WT, blue for Alpha, orange for Beta, green for Epsilon, and gray for Delta). Interacting residues are depicted as the solid sticks and residues losing their interactions are shown as the transparent sticks. RBD and ACE2 are presented in light gray and yellow, respectively.
(A) The average number of contacts between RBD residue 501 and ACE2. (B and C) Representative snapshots at D = 53 Å of (B) Alpha variant and (C) WT. (D) The average number of contacts between RBD residue 417 and ACE2 and (E and F) their interacting residue pairs at D = 53 Å of (E) Beta and (F) Alpha variants. (G) The average number of contacts between RBD residue 478 and ACE2 and (H and I) key interaction pairs at D = 78 Å of (H) Delta and (I) Epsilon variants. The overall color scheme is the same as in Figure 1, and each mutated residue in each variant is shown using the same colors (i.e., red for WT, blue for Alpha, orange for Beta, green for Epsilon, and gray for Delta). Interacting residues are depicted as the solid sticks and residues losing their interactions are shown as the transparent sticks. RBD and ACE2 are presented in light gray and yellow, respectively.
The Epsilon variant
The Epsilon variant showed lower pulling forces, due to its having the lowest number of contacts with the ACE2. Whereas the K353 residue in most variants interacts with Q493, Q496, Q498, T500, N/Y501, G502, and Y505 in the ACE2, this is not the case with the Epsilon K353 residue. Instead, the number of contacts was lost by half or more.
The reason for this loss of contact was traced to the L452R mutation, which shows higher interaction with the neighboring L450 but less with L492. These residues are in separate beta-strands, which are shortened to half the original length by this mutation. The shortening of these residues ultimately destabilizes the RBD-ACE2 interface and makes it easy to pull them apart relative to the WT. This is borne out by the rapid decline in the prevalence of the Epsilon variant.
The L452R mutation is seen with the Kappa and Delta variants as well, but with an additional E484Q or T478K mutation, respectively. These have similar pulling force requirements to the WT, as they interact in the same way at ACE2 K353 and RBD Q493, Q496, Q498, T500, N/Y501, G502, and Y505, unlike the Epsilon RBD variant.
The Delta variant shows some unique characteristics, such as the T478K mutation, which is associated with the highest pulling force if the RBD-ACE2 complex is to be completely separated. Thus, the RBD of this variant makes a greater number of contacts with the ACE2 as compared to any other variant.
The K478 residue still has contact with ACE2 P84 and M82, unlike Epsilon. This may be due to the fact that the RBD first approaches the ACE2, thus allowing stronger interactions and, ultimately, stronger infectivity. This observation may explain why the Delta variant has quickly spread and become the dominant circulating strain of SARS-CoV-2 in many nations around the world.
Experimental assay
The researchers assayed protein binding experimentally using microscale thermophoresis (MST). This test can detect viral protein-receptor interactions. The experiment showed that the WT RBD and the ACE2 receptor had binding affinities of approximately 27.5 ± 4.8 nanomolar (nM), which confirms earlier surface plasmon resonance.
The binding affinity of the Alpha variant for the ACE2 receptor was more than doubled relative to that of the WT; however, the Beta and Delta variants had a 20-30% greater affinity. The Epsilon variant had a 15% lower affinity as compared to the WT.
Study takeaways
The current study finds that the Alpha requires needs the highest pulling force to separate the RBD from the ACE2 receptor. The Beta, Gamma, and Delta variants follow these force profiles, which is likely due to the K417N/T mutations in the beta and Gamma lineages.
The Epsilon is easily dissociated due to the L452R mutation that reduces the stability of the RBD. Conversely, the Delta variant interacts most strongly with the receptor over a longer distance from the RBD-ACE2 complex. The study, therefore, provides important data on understanding the differences between the variants and how they interact with the receptor.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Kim, S. Liu, Y., Lei, Z., et al. (2021). Differential Interactions Between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. bioRxiv. doi:10.1101/2021.07.23.453598. https://www.biorxiv.org/content/10.1101/2021.07.23.453598v1.
- Peer reviewed and published scientific report.
Kim, Seonghan, Yi Liu, Zewei Lei, Jeffrey Dicker, Yiwei Cao, X. Frank Zhang, and Wonpil Im. 2021. “Differential Interactions between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern.” Journal of Chemical Theory and Computation 17 (12): 7972–79. https://doi.org/10.1021/acs.jctc.1c00965. https://pubs.acs.org/doi/10.1021/acs.jctc.1c00965.