Researchers in the United States have provided important insights into the antigen-specific cellular response and evolving humoral (antibody) response to messenger RNA- (mRNA) based vaccination against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
Using single-cell profiling technologies and machine learning analyses, the team identified and characterized antigen-specific T cell, B cell and antibody responses to Pfizer-BioNTech’s mRNA-based BNT162b2 vaccine in a longitudinal cohort of healthy donors.
“These cell and antibody associations may drive further efforts to predict vaccine effectiveness and identify mechanisms of protection,” writes Jonathan Irish from Vanderbilt University Medical Center in Nashville and colleagues.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
More about mRNA-based COVID-19 vaccines
Since the COVID-19 outbreak first began in late December 2019, intense research efforts have led to the emergency use authorization and mass roll-out of several mRNA-based vaccines designed to protect against infection with SARS-CoV-2.
These vaccines encode the SARS-CoV-2 spike protein – the main structure the virus uses to infect cells and a primary target of neutralizing antibodies following natural infection.
“These vaccines introduce the minimal genetic information to express viral antigens of interest and mimic natural infection of RNA viruses, such as SARS-CoV-2,” writes the team.
The challenges faced in understanding antigen-specific responses
While several research groups have identified increased T cell, myeloid and antibody responses to these vaccines, the cellular mechanisms that drive antigen-specific responses remain poorly understood.
“It is also not well established how pre-existing immunity to endemic coronaviruses impacts the B cell and antibody response against SARS-CoV-2 vaccination and how the antibody repertoire may evolve over time,” says the team.
While the reliable identification of antigen-specific cells poses a significant challenge in studies of immune responses to emerging diseases, Irish and colleagues say that single-cell machine learning analysis tools such as the Tracking Responders EXpanding (T-REX) algorithm and Linking B cell Receptor to Antigen specificity through sequencing (LIBRA-seq) combined with whole transcriptome RNA sequencing may help to address this need.
“These single-cell approaches identify rare cells that specifically expand following vaccination or infection that can be overlooked when analyzing cellular populations in bulk,” they write.
What did the researchers do?
The team used single-cell technologies to track the development of antigen-specific cellular and antibody responses to Pfizer-BioNTech’s BNT162b2 vaccine in longitudinal samples taken from a cohort of healthy donors, including one with breakthrough SARS-CoV-2 infection.
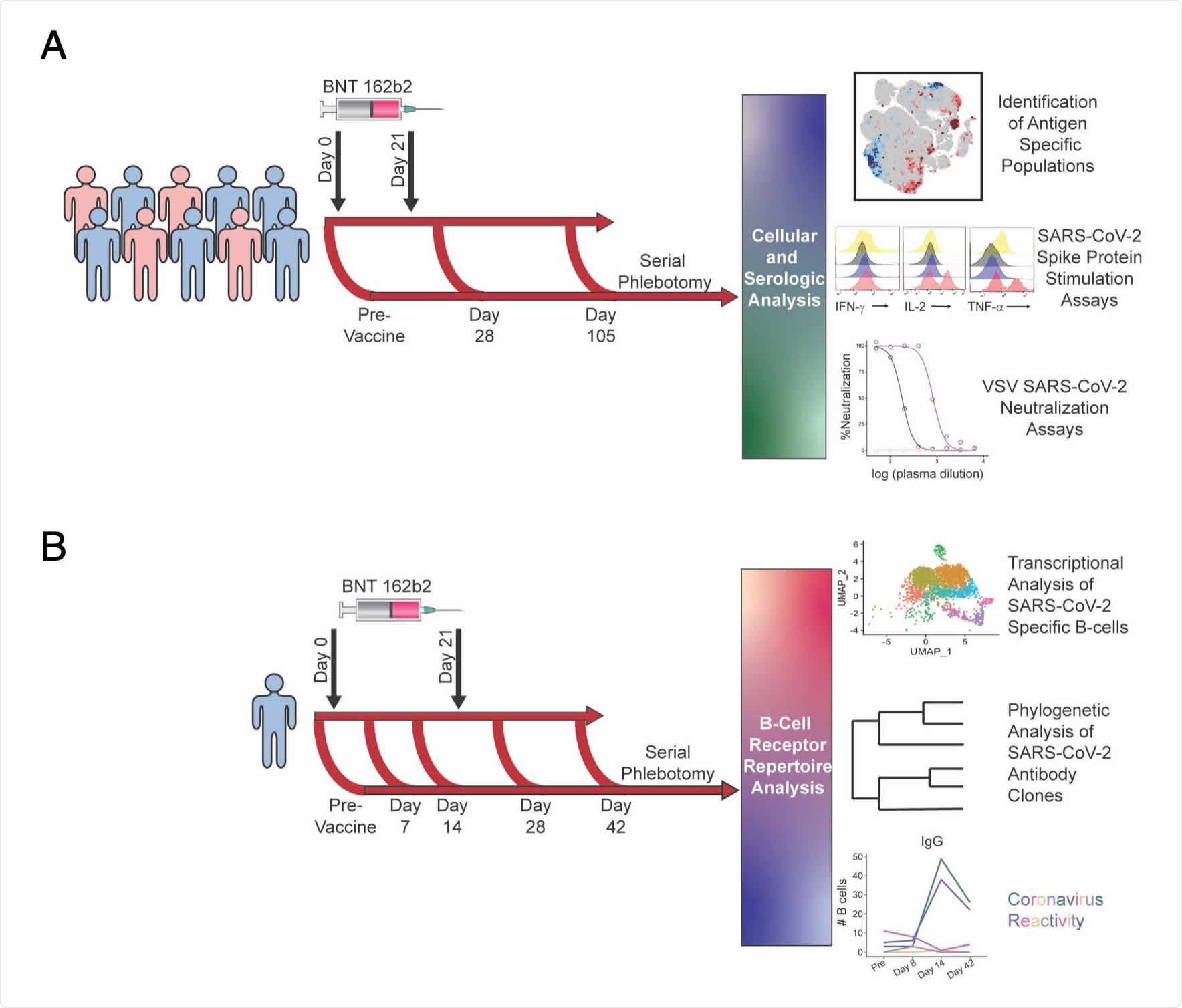
Schematic of vaccination schedule and sample collection. All donors were vaccinated with BNT162b2 on days 0 and 21. A. 10 healthy donors underwent serial phlebotomy was performed pre-vaccination (day -3 to 0), on day 28-30, and on day 105- 108. PBMCs were isolated at each time point, and citrated plasma was stored when possible. PBMCs from these donors were utilized for CyTOF and in vitro stimulation studies. Plasma was used both for SARS-CoV-2 ELISAs and vesicular stomatitis virus pseudoneutralization assays. B. A single healthy donor underwent serial phlebotomy prevaccination and on days 7, 14, 28, and 42. PBMCs and citrated plasma were isolated at each time point and used for transcriptional analysis of SARS-CoV-2 specific B cells.
T-REX pinpointed novel expanding populations of spike protein-specific non-canonical memory CD4+ and CD8+ T cells following vaccination.
“The approach allowed isolation of live, virus-specific T cells that will facilitate further studies,” says Irish and colleagues.
Analysis using LIBRA-seq identified both SARS-CoV-2-specific B cells and B cells that were cross-reactive between SARS-CoV-2 and other coronaviruses.
The B-cell repertoire shifted from apparent cross-reactivity to endemic coronaviruses prior to vaccination, to a more SARS-CoV-2-specific response that was marked by an expansion of immunoglobulin A (IgA) and IgG memory B cells and plasmablasts.
Donor deficient in T cell subsets developed breakthrough infection
Importantly, the antigen-specific cell subsets identified here correlated with a long-lasting IgG response that was lacking in the donor who developed a breakthrough infection. The donor was deficient in both the CD4 and CD8 populations of ICOS+CD38+ T cells.
Previous studies have shown that CD8+ T cells support life-long immunity against viruses such as influenza, Epstein-Barr and cytomegalovirus. The induction of a robust CD8+ T cell response is an emerging focus in vaccine development.
“Interestingly, cellular immunity may provide protection to individuals who mounted a suboptimal humoral antibody response and the donor with breakthrough SARS-CoV-2 infection failed to generate CD8+CD38+ICOS+ T cells,” says Irish and colleagues.
“If confirmed in a larger cohort, CD8+CD38+ICOS+ T cells may provide a novel marker of successful immunization and protection following SARS-CoV-2 vaccination,” they add.
What are the implications of the study?
The researchers say that the multi-compartment response induced by the BNT162b2 vaccine may have important ramifications, particularly among immunocompromised individuals.
Reduced humoral responses to the vaccine have already been reported in patients with solid organ transplant, cancer, or immune-mediated inflammatory diseases.
The identification of altered proteins or specific cell populations that could serve as biomarkers for successful vaccination may provide important insights for monitoring the development and continued protection within these vulnerable populations.
“While limited sample sizes here reduce statistical power for population-level correlations and detailed outcomes, the unbiased identification of antigen-specific CD4, CD8, and B cell populations provides important insight into general mechanisms of RNA-based vaccines and the cellular basis for vaccine-induced antibodies and protection from SARS-CoV-2,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Irish JM, et al. Single-Cell Profiling of the Antigen-Specific Response to BNT162b2 SARS-CoV-2 RNA Vaccine. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.07.28.453981, https://www.biorxiv.org/content/10.1101/2021.07.28.453981v1
- Peer reviewed and published scientific report.
Kramer, Kevin J., Erin M. Wilfong, Kelsey Voss, Sierra M. Barone, Andrea R. Shiakolas, Nagarajan Raju, Caroline E. Roe, et al. 2022. “Single-Cell Profiling of the Antigen-Specific Response to BNT162b2 SARS-CoV-2 RNA Vaccine.” Nature Communications 13 (1): 3466. https://doi.org/10.1038/s41467-022-31142-5. https://www.nature.com/articles/s41467-022-31142-5.