Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the earlier SARS-CoV-1 gain entry to host cells by interacting with the ACE2 receptor, while the closely related MERS-CoV enters human cells by binding with dipeptidyl-peptidase.
Besides these major receptors each virus also interacts with a number of additional surface proteins or glycans that aid in initial adhesion and subsequent internalization.
SARS-CoV-2, for example, is known to involve cell surface protease TMPRSS2 to cleave a section of its own spike protein to facilitate cell entry, and a number of other potential receptors have recently been described, including kringle containing transmembrane protein 1 and asialoglycoprotein receptor 1.
Similarly, MERS-CoV is known to interact with some sialic acids, sugar molecules found on the surface of cells, which have also been suggested as a candidate SARS-CoV-2 receptor. Sialic acids are usually found in the terminal position on a variety of glycoconjugates, on the surface of a wide variety of cells, and are also known to mediate the attachment of other viruses such as influenza.
In a paper recently uploaded to the journal Frontiers in Chemistry by Li et al. (July 22nd, 2021) the interaction between the SARS-CoV-2 spike protein and N-acetylneuraminic acid (NANA), the most commonly found sialic acid in humans, is investigated, finding that the molecule may act as a secondary or supplementary receptor to the virus.
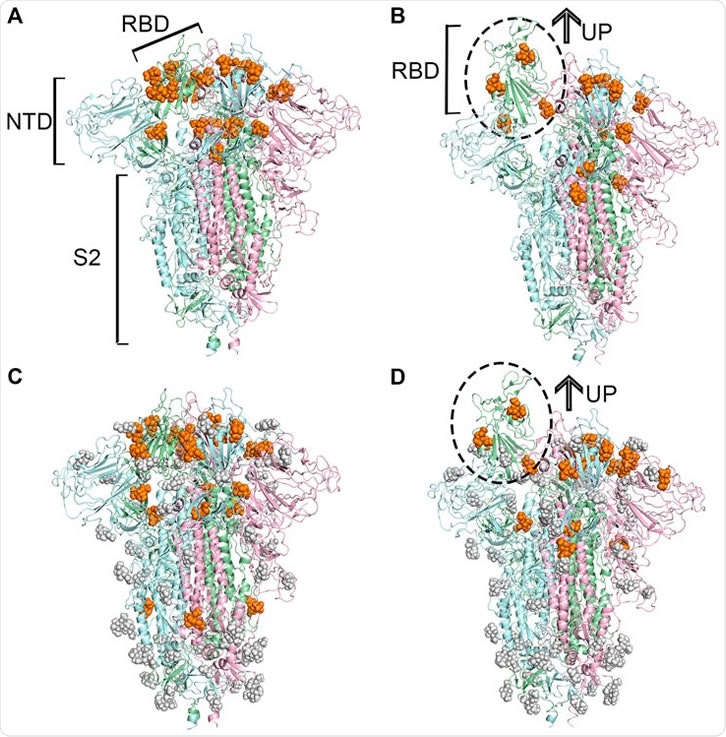
Predicted potential bindings of sialic acids on the surface of the spike protein. Potential ligandable binding sites were identified on the four constructed models of the trimeric spike protein by using FTMap (Ngan et al., 2012) and SiteMap (Halgren, 2009). The different colors of the cartoon models in each figure represent different chains of the protein: chain A is shown in pale-green, chain B is in pale-blue, and chain C is in light-pink. The protein structures which contain gray sphere balls represent glycosylated state S trimer (the gray sphere models are the glycosylation), and the orange balls represent sialic acid molecules. (A) 21 and (B) 15 sialic acid molecules were observed to bind to the different places of the surface of unglycosylated spike protein in “down” and “up” conformational states, respectively, (C) 23 potential sites for sialic acid binding were identified on the surface of glycosylated spike protein in “down” conformational state, and 17 were found (D) in the “up” conformational state.
How was the study performed?
The SARS-CoV-2 spike protein receptor-binding domain (RBD) was modeled computationally, in both open and closed conformations and with and without glycosylation, and each examined for potential binding sites for NANA. When in the closed state, the RBD bore 21 potential binding sites when non-glycosylated and 23 when glycosylated, while the open state RBD had 15 and 17, respectively, totaling 40 unique sites.
The stability of each binding site was assessed in 20 nanosecond long simulations, finding overall that glycosylation was beneficial to bond stability. Longer 200 ns simulations were performed for the best candidates, finding a number of appealing sites.
The group notes that while in the closed conformation, an additional binding site that spans the RBD is available, being unsuitable when in the open conformation.
The major binding site of sialic acids to MERS-CoV is within the N-terminal domain of the virus's spike protein, while in the case of SARS-CoV-2, the group detected no binding sites here, only within the RBD.
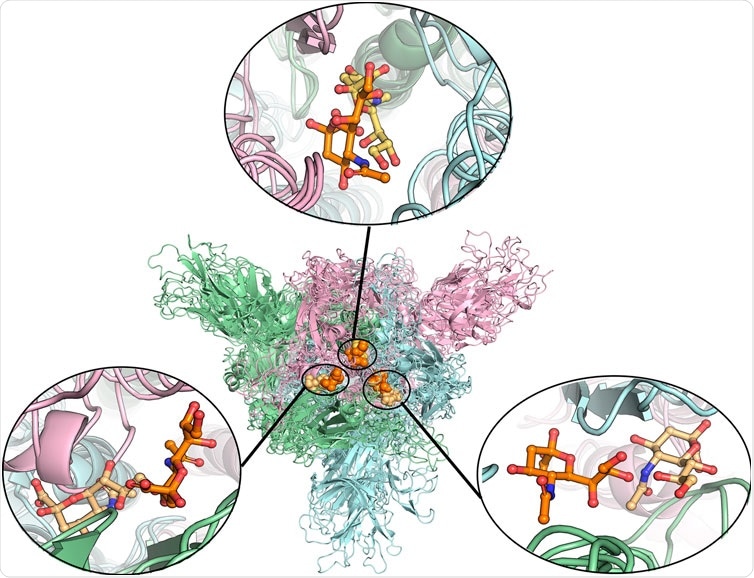
Alignment of different protein structures to identify the overlapped sialic acid-binding positions, that is, SA_6, SA_7, and SA_24. The figure shows the alignment of these final stable complex structures of sialic acid with spike protein, generated by the MD simulations starting from those four different protein structures. The sialic acid-binding positions, i.e., SA_6, SA_7, and SA_24 were found to be conserved in “down” conformational states of the spike protein and to be observable in “up” conformational states. Position SA_6 is between chain B (pale-blue) and chain C (light-pink), position SA_7 is between chain A (pale-green) and chain B and position SA_24 is between chain A and chain C. The stable bound sialic acids are shown in different colors. Three sialic acids in the glycosylated “down” conformational state are shown in orange color. Two sialic acids in the unglycosylated “down” conformational state are shown in light orange color. One sialic acid bound on the glycosylated “up” conformational state is shown in yellow-orange color. The detailed interaction modes for the bound modes are shown in Figure 4 and Supplementary Figures S7, S8. No stable sialic acid appears in these three positions for the glycated “up” state.
Once bound to a host cell, the spike protein of SARS-CoV-2 undergoes several conformational changes that allow it to pass the cell membrane, some of which are mediated by molecules binding to the protein at particular sites.
The group suggests that the examined sialic acid may potentially be involved in inducing such changes, as one of the major binding pockets identified overlaps with one thought to be involved in conformational changes. However, this has yet to be confirmed.
The identification of this binding site may facilitate drug design efforts, wherein a drug with a similar structure or with a sialic acid targeting group could potentially interfere with the SARS-CoV-2 cell entry process.
Journal reference:
- Li Bingqian, Wang Lin, Ge Huan, Zhang Xianglei, Ren Penxuan, Guo Yu, Chen Wuyan, Li Jie, Zhu Wei, Chen Wenzhang, Zhu Lili, Bai Fang, Identification of Potential Binding Sites of Sialic Acids on the RBD Domain of SARS-CoV-2 Spike Protein, Frontiers in Chemistry, DOI=10.3389/fchem.2021.659764, https://www.frontiersin.org/article/10.3389/fchem.2021.659764