Among all the coronaviruses that can infect humans, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), has been recognized as the most infectious. Other coronaviruses that have caused epidemics include the Middle East respiratory syndrome (MERS-CoV) and the severe acute respiratory syndrome coronavirus (SARS-CoV). Typically, these viruses infect the respiratory tracts and cause pneumonia-like symptoms.
The novel SARS-CoV-2 was first reported in Wuhan, China, in December 2019. Subsequently, owing to mutations, several SARS-CoV-2 variants such as the Alpha, Beta, Delta and Gamma variants have emerged.
The genomic sequence of SARS-CoV-2 has led to the identification of four structural proteins including the spike (S), membrane (M), envelop (E), and nucleocapsid (N) proteins, as well as other non-structural and accessory proteins.
All of the COVID-19 vaccines that have received emergency use authorization (EUA) from various federal regulatory bodies throughout the world are based on the S protein due to its crucial role in SARS-CoV-2 infection.
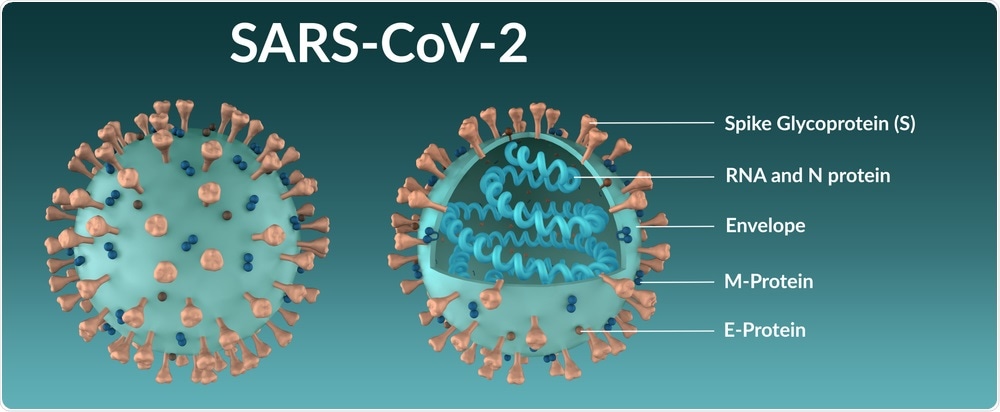
How does SARS-CoV-2 infect cells?
The SARS-CoV-2 protein mediates viral entry inside the host cell. The S protein contains two domains that include S1 and S2.
The S1 domain consists of receptor-binding domains (RBDs) that directly bind with the angiotensin-converting enzyme 2 (ACE2) receptor, which is present on the host’s cell surface. Comparably, the S2 domain contains the fusion peptide that allows for the fusion of SARS-CoV-2 to the cell membranes.
Several changes occur at the molecular level following viral invasion to allow for this virus to control the host’s cellular mechanisms and initiate viral replication. As the binding of RBDs to ACE2 plays a key role in viral pathogenesis, inhibition of this binding could prevent viral infection.
Traditional medicinal plants and viral infection
Plant extracts containing different secondary metabolites possess various medicinal properties, such as antimicrobial and antispasmodic activity, with minimal side effects. For example, thyme was once used for the treatment of asthma, laryngitis, the common cold, and throat congestion. During the sixteenth and seventeenth centuries, wild thyme was used for the treatment of malaria and epilepsy. The Balkans also used thyme as anticholesterolemic and immunostimulant.
Another interesting immunomodulatory property of thymol is that it can enhance the expression of CD4, CD8, and Th1 cytokines through upregulation of interferon 4 (IFN-4) expression. This agent can also boost the secretion of interleukin-12 (IL-12).
Emodin is another phytocompound that acts as an inhibitor of the 3a ion channel of SARS-CoV and HCoV-OC43 coronaviruses. Previous studies have also revealed that high concentrations of emodin can inhibit different stages of the virus life cycle.
A new study
Considering the antiviral activity of medicinal herbs, a new study published in the journal Current Pharmacology Reports focuses on the identification of potent phytochemicals from Himalayan herbs, namely, Rheum emodi, Thymus serpyllum, and Artemisia annua, for the treatment of COVID-19.
Herein, an in silico approach demonstrated the potential of unique phytocompounds (emodin, thymol, and carvacrol) to bind SARS-CoV-2 S glycoproteins (6VXX and 6VYB), the B.1.351 variant’s S glycoprotein, and also with ACE2. These phytocompounds could also prevent the binding of SARS-CoV-2 to the host’s ACE2, TMPRSS2, and neutrapilin-1 receptors.
Chloroquine has previously been used for the treatment of COVID-19. Owing to this, the authors compared the binding pattern of chloroquine and artemisinin with S proteins (6VXX, 6VYB) and its variant 7NXA, to understand its mode of action and inhibitory effects.
In this study, researchers used the AutoDock/Vina software to identify phytochemicals that can bind with the S protein. These molecular docking studies screened numerous phytocompounds and narrowed them down to emodin, which showed the best binding affinity with 6VYB and SARS-CoV-2 B.1.351 variant (7NXA). Emodin also showed the best interactions with TMPRSS 2 and ACE2.
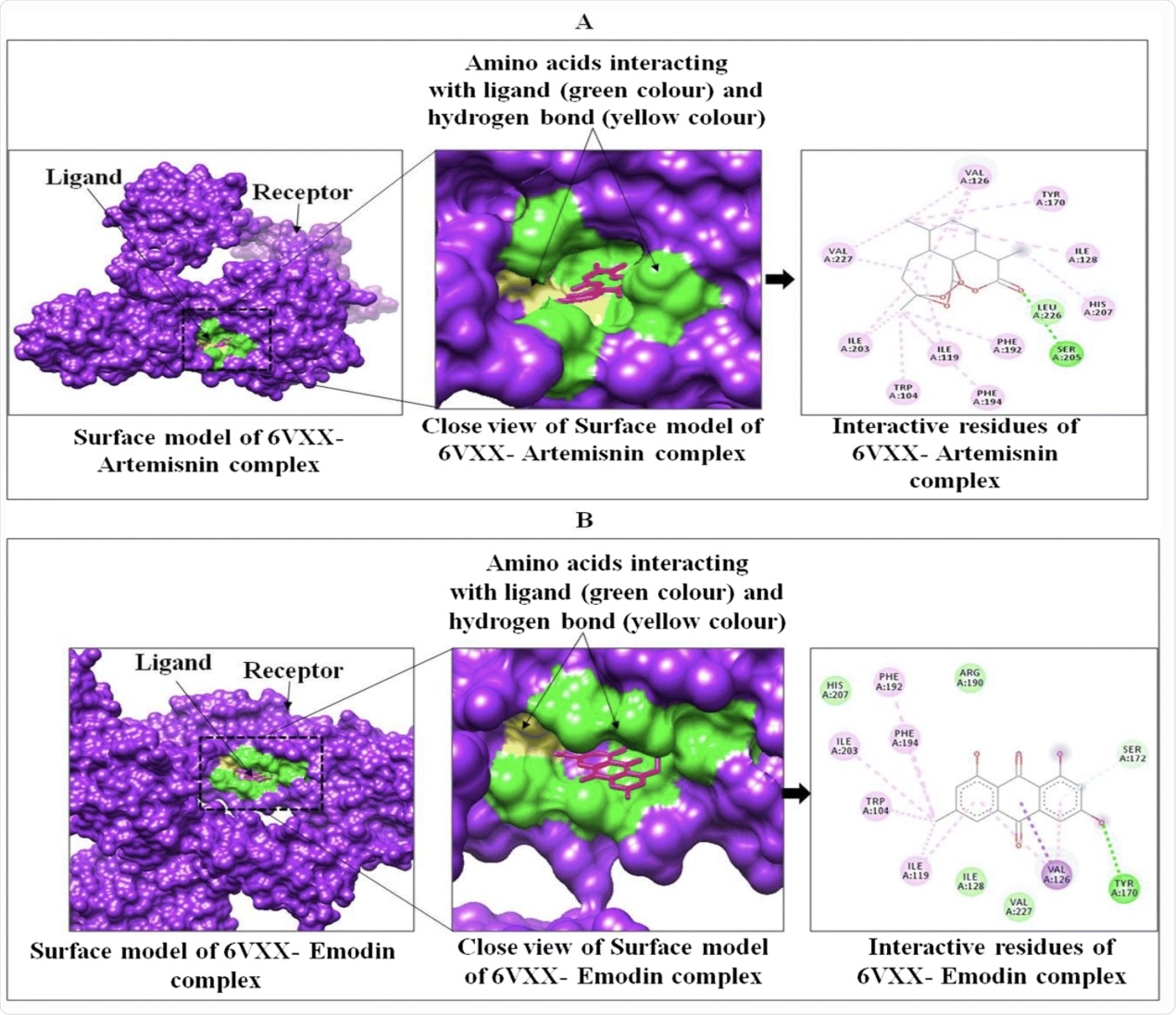
Interactions between targeted protein receptor SARS-CoV-2 spike glycoprotein (PDB ID: 6VXX) with artemisinin (A) and emodin (B) using Chimera.
In this study, although the lowest binding energy was observed in artemisinin with 6VXX and 6VYB, this compound showed significant interaction with TMPRSS 2 and ACE2. Molecular dynamics (MD) simulation was also performed for 50ns. In the Ramachandran plot developed after MD, both the S-protein domains showed less than 1.0% outlier amino acid residues.
Taken together, the MD simulation study revealed that the interaction of artemisinin with 6VYB as compared to 6VXX was more stable. The researchers also found that all phytocompounds that were evaluated in this study are non-toxic and non-carcinogenic.
Conclusion
Considering these results, the authors recommend artemisinin as a potential phytocompound that can effectively inhibit the interaction between SARS-CoV-2 and ACE-2 receptor, especially when the S protein is in an open state conformation.
The current research is unique because it has compared the open (6YVB) and closed (6VXX) conformations of S proteins. Interestingly, the open state of the S protein (6YVB), which was found to be more pathogenic, showed more stable interaction with artemisinin as compared to the closed state (6VXX).
Future in vitro and in vivo studies will need to be conducted to determine the therapeutic efficacy of artemisinin for the treatment of COVID-19.