The introduction of nucleosides to inhibit the RNA-dependent RNA polymerase (RdRp) enzyme is one important mechanism of antiviral drugs such as remdesivir; however, the presence of ExoN renders these drugs useless. Therefore, understating the structure of ExoN could help identify or assist in the development of inhibitory molecules to ultimately accelerate drug discovery processes.
 Study: Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Image Credit: creativeneko / Shutterstock.com
Study: Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Image Credit: creativeneko / Shutterstock.com
Background
To date, there are no effective antivirals against SARS-CoV-2. However, its replication/transcription complex (RTC) is a promising target, along with its core of RdRp and other non-structural proteins (nsps). Nucleotide analogs like remdesivir insert the wrong nucleotide into the SARS-CoV-2 genome in order to arrest replication; however, this event can be rescued by the proofreading activity of ExoN.
This enzyme, which is found as the N-terminal ExoN domain of the viral non-structural protein 14 (nsp14), is stimulated by nsp10, which also stabilizes its active site structure. ExoN also breaks down viral double-stranded RNA (dsRNA), which would otherwise activate host-pathogen recognition receptors (PRRs). This contributes to the immune escape capabilities of SARS-CoV-2.
The current study discusses the molecular binding of substrates by ExoN, as well as how this enzyme recognizes and removes wrongly inserted nucleotides or analogs in the newly forming RNA strand.
Study details
The scientists used a hairpin RNA substrate known as TSP31 to explore the catalytic activity of ExoN. This allowed binding to RNA but not RNA cleavage by the enzyme.
Using a combination of size-exclusion chromatography (SEC), single-particle cryo-electron microscopy (EM), and in silico methods, the researchers found that the nsp10-nsp14-RNA complex binds also to nsp8, though in a weak and dynamic manner. This binding promotes stable RNA substrate binding that allows for RNA cleavage by ExoN. It also increases RNA degradation by the nsp10-nsp14 complex.
As a common factor to both RdRp and ExoN enzyme complexes, nsp8 appears to play a crucial role in transferring the RNA substrate between these molecules. However, more work will need to be conducted to determine how nsp8 corrects mismatched nucleotides.
When a mutant form of the ExoN was studied, part of the sample contained the tetramerized form of the nsp10-nsp14-RNA complex, which appears to prevent nsp8 binding.
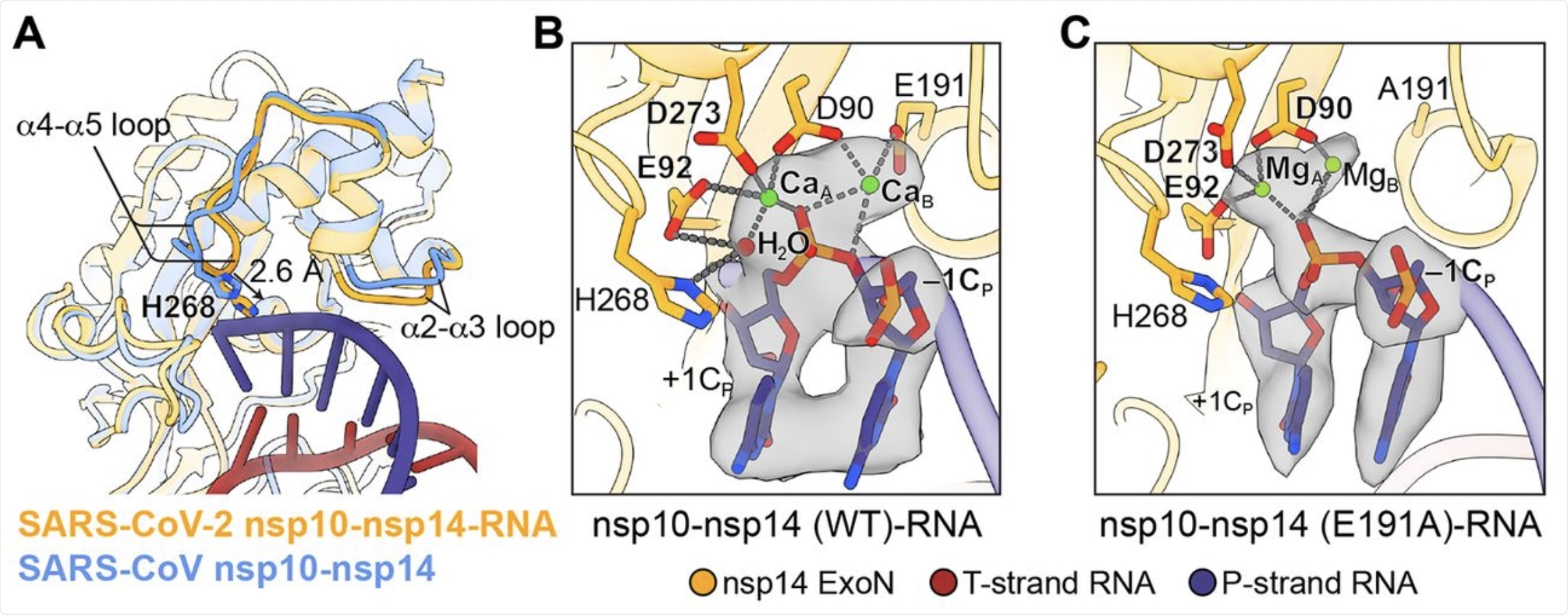 (A) Superimposition of SARS-CoV nsp10-nsp14 complex (cornflower blue, PDB 5C8U) and SARS-CoV-2 nsp10-nsp14-RNA complex (orange) illustrates the conformational changes of α2-α3 and α4-α5 loops and a 2.6 Å shift of H268 toward the RNA upon substrate binding. (B) Active site structure of SARS-CoV-2 nsp10-nsp14 (WT)-RNA complex. Ca2+ ions, green spheres; catalytic water, red sphere. Nucleotide residues in P-strand RNA are indicated with subscript “P.” +1CP, –1CP, the catalytic water and two active site metal ions are superimposed with their cryo-EM densities contoured at 10σ. (C) Active site structure of SARS-CoV-2 nsp10-nsp14 (E191A)-RNA complex. Mg2+ ions, green spheres. +1CP, –1CP, and two active site metal ions are superimposed with their cryo-EM densities contoured at 7σ.
(A) Superimposition of SARS-CoV nsp10-nsp14 complex (cornflower blue, PDB 5C8U) and SARS-CoV-2 nsp10-nsp14-RNA complex (orange) illustrates the conformational changes of α2-α3 and α4-α5 loops and a 2.6 Å shift of H268 toward the RNA upon substrate binding. (B) Active site structure of SARS-CoV-2 nsp10-nsp14 (WT)-RNA complex. Ca2+ ions, green spheres; catalytic water, red sphere. Nucleotide residues in P-strand RNA are indicated with subscript “P.” +1CP, –1CP, the catalytic water and two active site metal ions are superimposed with their cryo-EM densities contoured at 10σ. (C) Active site structure of SARS-CoV-2 nsp10-nsp14 (E191A)-RNA complex. Mg2+ ions, green spheres. +1CP, –1CP, and two active site metal ions are superimposed with their cryo-EM densities contoured at 7σ.
Mechanism of RNA substrate binding
The investigators found that the binding of the RNA substrate to the ExoN active site of the SARS-CoV-2 wildtype (WT) nsp10-nsp14-RNA complex causes two metal ions to bind to the catalytic center. This triggers further reactions that complete the active site.
The detailed description of the interactions with the dsRNA suggests that the enzyme maintains its specificity for the substrate. Furthermore, these details provide information on the structure of the substrate relative to other RNA viral and proofreading ExoNs, as well as the predicted SARS-CoV ExoN.
Importantly, all coronavirus ExoNs share the same RNA-contacting residues in nsp14. Thus, they have the same substrate recognition mechanism.
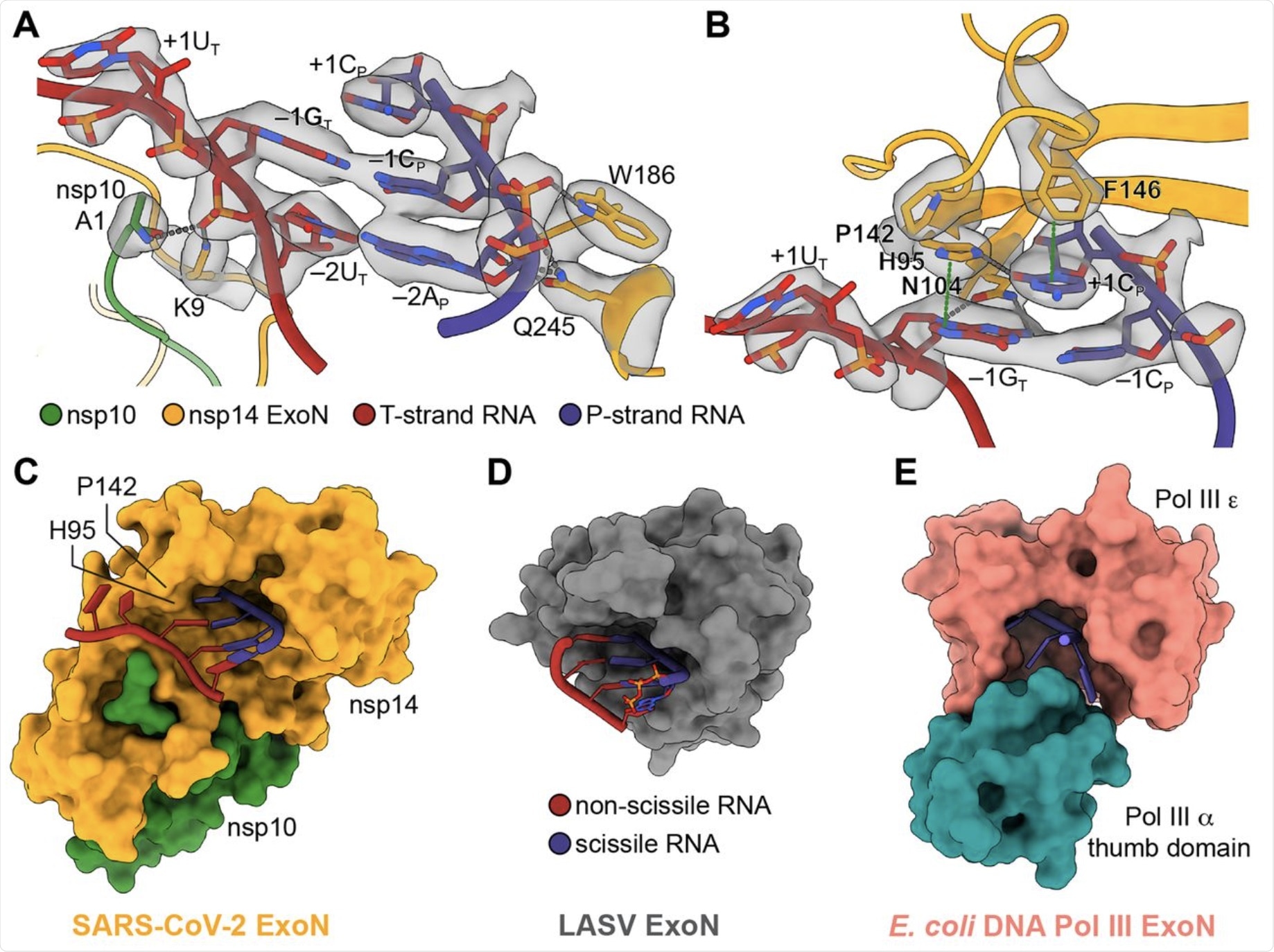 (A) Interactions between SARS-CoV-2 nsp10-nsp14 ExoN and T35P31 RNA backbone. Nucleotide residues in T-strand RNA are indicated with subscript “T,” nucleotide residues in P-strand RNA are indicated with subscript “P.” Hydrogen bonds and salt bridges are shown as gray dotted lines. Interacting nucleotide and protein residues are superimposed with their cryo-EM densities contoured at 7σ. (B) Interactions between SARS-CoV-2 nsp10-nsp14 ExoN and T35P31 RNA at +1 and –1 nucleobases positions. Hydrogen bonds and salt bridges are shown as gray dotted lines. π-π stacking interactions are indicated by green dotted lines. Interacting nucleotide and protein residues are superimposed with their cryo-EM densities contoured at 7σ. (C) Surface representation of SARS-CoV-2 nsp10-nsp14 ExoN substrate-binding pocket shows a restricted opening on the T-strand side that prevents base-pairing at substrate RNA +1 position. For clarity, the N7-MTase domain of nsp14 is not shown. (D) Surface representation of LASV NP ExoN domain. A fully base-paired dsRNA substrate (shown as cartoons) is bound in the substrate-binding pocket of LASV ExoN. (E) Surface representation of E. coli DNA Pol III ExoN complex. The narrow substrate-binding pocket allows the entry of ssDNA only.
(A) Interactions between SARS-CoV-2 nsp10-nsp14 ExoN and T35P31 RNA backbone. Nucleotide residues in T-strand RNA are indicated with subscript “T,” nucleotide residues in P-strand RNA are indicated with subscript “P.” Hydrogen bonds and salt bridges are shown as gray dotted lines. Interacting nucleotide and protein residues are superimposed with their cryo-EM densities contoured at 7σ. (B) Interactions between SARS-CoV-2 nsp10-nsp14 ExoN and T35P31 RNA at +1 and –1 nucleobases positions. Hydrogen bonds and salt bridges are shown as gray dotted lines. π-π stacking interactions are indicated by green dotted lines. Interacting nucleotide and protein residues are superimposed with their cryo-EM densities contoured at 7σ. (C) Surface representation of SARS-CoV-2 nsp10-nsp14 ExoN substrate-binding pocket shows a restricted opening on the T-strand side that prevents base-pairing at substrate RNA +1 position. For clarity, the N7-MTase domain of nsp14 is not shown. (D) Surface representation of LASV NP ExoN domain. A fully base-paired dsRNA substrate (shown as cartoons) is bound in the substrate-binding pocket of LASV ExoN. (E) Surface representation of E. coli DNA Pol III ExoN complex. The narrow substrate-binding pocket allows the entry of ssDNA only.
Other findings
The researchers found that a free 3′-OH of the RNA substrate is essential for ExoN-mediated breakdown of RNA. This suggests that 3′-deoxy nucleotide analogs can potentially act as effective RdRp chain terminators while being resistant to ExoN excision.
The SARS-CoV-2 ExoN was also found to accept both single-stranded RNA (ssRNA) and dsRNA substrates, thereby indicating that mismatch correction in vivo may operate in at least two different ways.
The authors also observed that RNA containing remdesivir monophosphate (RMP), following the incorporation of the inhibitor, could still be bound by the viral ExoN. This agrees with the finding that this RMP-terminated RNA remains vulnerable to the enzyme, while the absence of the ExonN increases its susceptibility. Other modifications should also be explored.
What are the implications?
This study discusses how mismatch correction occurs during RNA synthesis by the SARS-CoV-2. The authors also elucidate which parts of the enzyme architecture are required for its recognition of the RNA substrate and catalytic action.
Taken together, the authors offer several suggestions on possible alterations that could be made on the ribose structure to prevent ExoN-mediated resistance to nucleotide analog RdRp inhibitors. This information could be particularly useful during the design of powerful ExoN inhibitors that can be combined with the drugs like remdesivir for more effective action against SARS-CoV-2.
“Understanding this structure and the molecular details of how ExoN works can help guide further development of antivirals.”