A recent study by Canadian and US researchers demonstrates how frameshift signals of coronaviruses represent a viable target for developing novel therapeutics with broad-spectrum activity – not only against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) but also other constituents of the coronavirus family. The study is currently available on the bioRxiv* preprint server while it undergoes peer review.
Recurring outbreaks of novel zoonotic coronavirus diseases since 2000, especially the ongoing coronavirus disease (COVID-19) pandemic caused by SARS-CoV-2, have underscored the significance of developing therapeutics that have broad-spectrum activity against coronaviruses.
A research community pinpointed one event in molecular genetics known as programmed ribosomal frameshifting as a possible solution to this problem. This represents an alternate mechanism of translation that merges proteins encoded by two overlapping open reading frames.
Consequently, as all coronaviruses use –1 programmed ribosomal frameshifting (–1 PRF) in order to handle the expression of key viral proteins, the frameshift signal in viral messenger RNA (mRNA) that stimulates –1 PRF, represents a promising potential target for broad therapeutics.
Thus far, several studies have identified small-molecule ligands with the propensity to modulate –1 PRF in human coronaviruses. More recently, empirical screens in SARS-CoV-2 for modulation have unveiled a number of compounds that either suppress or enhance –1 PRF.
Hence, in order to test the viability of this strategy, a research group led by Dr. Sneha Munshi from the University of Alberta in Edmonton (Canada) decided to explore a group of six small-molecule ligands by evaluating their activity against the frameshift signals from representative bat coronaviruses (i.e., the most probable source of future zoonoses), but also against SARS-CoV-2 and MERS-CoV.
Candidates in coronaviral sequences
By performing multiple sequence alignment of 959 bat coronaviral sequences from the NCBI virus database (a resource that provides viral genome sequence data and associated information), this research group has built a representative panel for their study. Of these sequences, 48 had a substantial coverage of the frameshift region.
Furthermore, by using dual-luciferase assays of frameshifting, they have initially confirmed the activity of the putative –1 PRF signals from a panel of four coronaviruses selected as representatives of the range of sequences found to date across bat coronaviruses.
Then they have augmented the pool of potential –1 PRF inhibitors by screening FDA-approved drugs for activity against –1 PRF in SARS-CoV-2. Finally, six small-molecule ligands were selected (based on prior studies) in order to compare their effects on –1 PRF induced by a panel of six coronaviral frameshift signals – four from bat coronaviruses, and two as from SARS-CoV-2 and MERS-CoV.
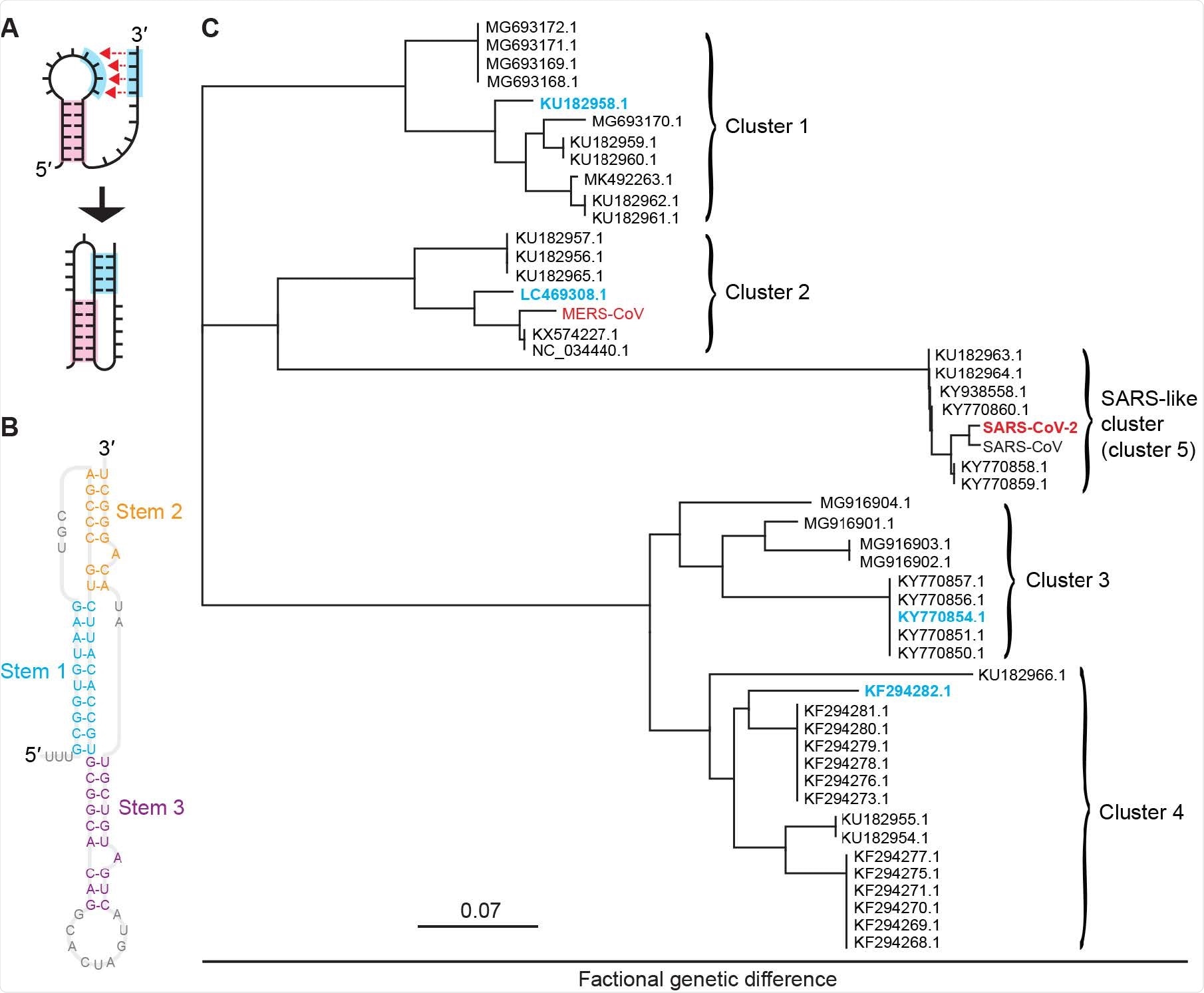
Pseudoknots from bat coronaviruses. (A) Pseudoknots form when the unpaired bases in an RNA stem-loop pair with another single-stranded segment. (B) CoV pseudoknots have a 3-stem architecture, illus-trated here for the pseudoknot from SARS-CoV-2. (C) Phylogenetic tree showing five clusters of bat and human CoVs with similar pseudoknot sequences. Representatives from each cluster studied here are shown in cyan for bat CoVs and red for human CoVs.
Nafamostat as the star candidate
In a nutshell, the researchers have observed that –1 PRF may be inhibited substantially for each of the frameshift signals from bat coronaviruses, suggesting that they can all be targeted therapeutically in a broad spectrum manner. However, the researchers have noted that the mechanism of action of the –1 PRF inhibitors is still not completely clear.
In any case, after examining the inhibitory effects of the tested compounds, the results have shown that –1 PRF can be moderate to strongly suppressed by at least one of the inhibitors for each of the representative frameshift signals in the testing panel.
And while some ligands had noteworthy activity against only a handful of the frameshift signals, the serine protease inhibitor known as nafamostat strongly suppressed –1 PRF in several of them – with limited or no effect on –1 PRF caused by frameshift signals from other viruses that have been used as negative controls.
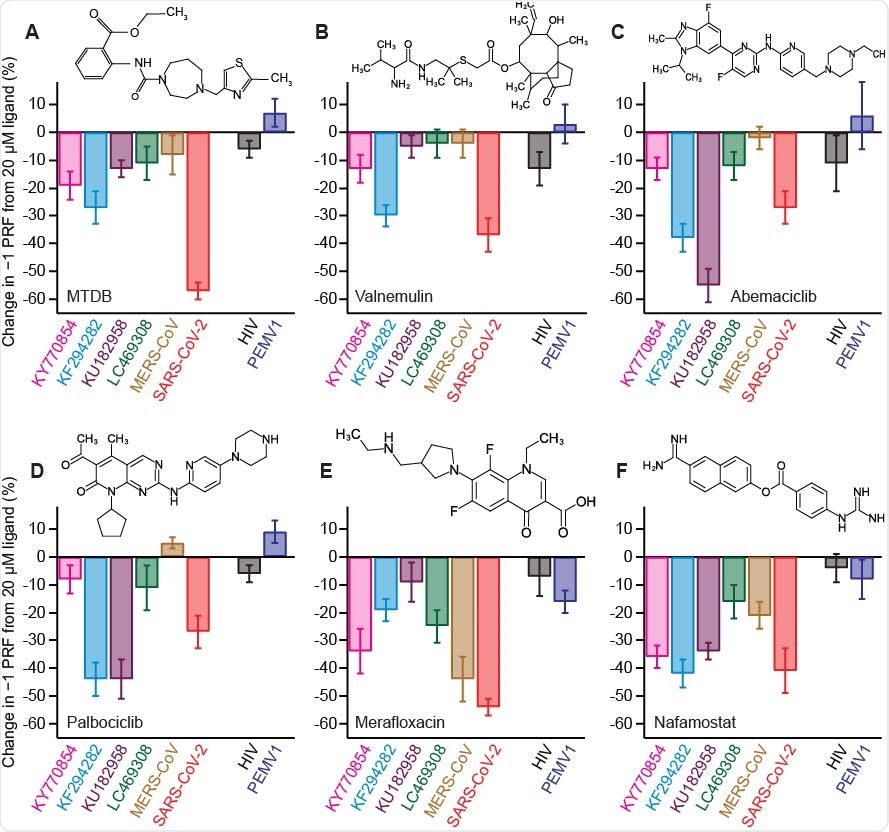
Activity of −1 PRF inhibitors against frameshift signals from different CoVs. (A) Change in −1 PRF efficiency compared to basal levels (Fig. 2) induced by 20 μM MTDB. Remaining panels show the same for (B) valnemulin, (C) abemaciclib, (D) palbociclib, (E) merafloxacin, and (F) nafamostat. In each case, results for CoVs are shown on left, results for specificity controls on right. Experiments performed in vitro using dual-luciferase reporter in rabbit reticulocyte lysate. Error bars represent s.e.m. from 3–7 replicates. Insets: chemical structures of inhibitors.
Far-reaching implications
These results imply the possibility of finding small-molecule ligands that inhibit –1 PRF specifically in a broad spectrum of coronaviruses. The compound nafamostat that was highlighted as the best candidate is, interestingly, already in clinical trials for treating COVID-19, primarily due to its recognized activity as a protease inhibitor.
“Indeed, there were no obvious patterns in terms of effectiveness against alpha coronaviruses versus beta coronaviruses for any of the inhibitors active against multiple frameshift signals,” say study authors in this bioRxiv paper.
“In every case, they were effective against at least one alpha coronavirus and one beta coronavirus, suggesting that the family to which the coronavirus belongs has little influence on the effectiveness of any of these inhibitors”, they add.
Hopefully, the results like these will prompt even more research endeavors on –1 PRF inhibitors in the future, as well as their interactions with frameshift signals and ribosomes. This will, in turn, enrich our treatment armamentarium not only against COVID-19 but potentially other emerging diseases caused by coronaviruses.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Munshi, S. et al. (2021). Small-molecule ligands can inhibit –1 programmed ribosomal frameshifting in a broad spectrum of coronaviruses. bioRxiv. https://doi.org/10.1101/2021.08.06.455424, https://www.biorxiv.org/content/10.1101/2021.08.06.455424v1
- Peer reviewed and published scientific report.
Munshi, Sneha, Krishna Neupane, Sandaru M. Ileperuma, Matthew T. J. Halma, Jamie A. Kelly, Clarissa F. Halpern, Jonathan D. Dinman, Sarah Loerch, and Michael T. Woodside. 2022. “Identifying Inhibitors of −1 Programmed Ribosomal Frameshifting in a Broad Spectrum of Coronaviruses.” Viruses 14 (2): 177. https://doi.org/10.3390/v14020177. https://www.mdpi.com/1999-4915/14/2/177.