A team of scientists from Frankfurt, Germany, has recently explored the efficacy of vaccine- or infection-induced antibodies and therapeutic monoclonal antibodies in neutralizing kappa and delta variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The findings reveal that the delta variant is more resistant to antibody-mediated neutralization than the kappa variant. The study is currently available on the medRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The immunogenic potency of SARS-CoV-2 can be altered by the emergence of mutations in major immunogenic components, including the spike protein. Since the current generation of coronavirus disease 2019 (COVID-19) vaccines and therapeutic antibodies have been developed against the original Wuhan strain, mutation-induced structural changes in the spike proteins of emerging variants can potentially alter their susceptibility to antibody-mediated neutralization.
Soon after its first detection in India during the second wave of the pandemic, the delta variant (B.1.617.2) of SARS-CoV-2 became dominant across the globe. Two spike mutations L452R and T478K, primarily account for significantly increased infectivity and immune evasion potency of the delta variant. Another subvariant of the B.1.617 lineage is kappa (B.1.617.1), which also originated in India. There is evidence claiming that, like the delta variant, the kappa variant with two spike mutations L452R and E484Q might be less susceptible to antibody-mediated neutralization.
In the current study, the scientists have explored the effectiveness of COVID-19 vaccines, convalescent sera, and monoclonal antibodies in neutralizing the kappa, delta, and epsilon variants of SARS-CoV-2. The epsilon variant (B.1.427/B.1.429) originated in California and contained L452R mutation in the spike protein.
Specifically, they have tested the virus-neutralizing efficacy of two mRNA-based COVID-19 vaccines, BNT162b2 (Pfizer/BioNTech) and mRNA1273 (Moderna), and three monoclonal therapeutic antibodies bamlanivimab, casirivimab, and imdevimab.
Virus neutralization by vaccinated and convalescent sera
As observed in the study, antibodies induced by vaccination or natural infection are less effective in neutralizing SARS-CoV-2 variants containing the L452R spike mutation.
Specifically, antibodies induced by natural infection showed considerably lower neutralizing potency against the kappa, delta, and epsilon variants compared to that against previously circulating SARS-CoV-2 variants with D614G spike mutation.
Similarly, vaccine-induced antibodies showed a 2-fold and 1-fold reduced ability to neutralize the kappa and delta variants and the epsilon variant, respectively. Compared to the epsilon variant, both kappa and delta variants showed 2-fold higher resistance to neutralization by vaccine- and infection-induced antibodies.
Virus neutralization by monoclonal antibodies
Among tested monoclonal antibodies, bamlanivimab failed to neutralize SARS-CoV-2 variants with L452R spike mutation. Compared to D614G-containing variants, the kappa variant showed a 5-fold lower susceptibility to casirivimab-mediated neutralization. However, neutralization of the kappa variant by imdevimab or a combination of casirivimab/imdevimab was almost comparable to that of previously circulating variants. Moreover, effective neutralization of the epsilon variant was observed in response to casirivimab or a combination of casirivimab/imdevimab.
In contrast to the kappa variant, the delta variant showed significantly low susceptibility to imdevimab and moderate susceptibility to casirivimab.
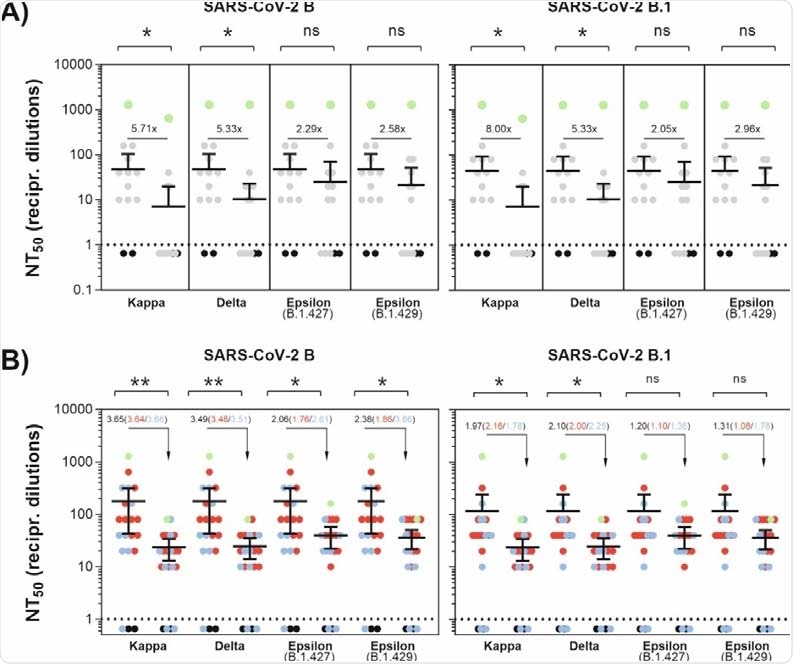
Antibody-mediated neutralization efficacy against SARS-CoV-2 variants. (A) A549-AT cells were incubated with serially diluted (1:2) sera from convalescent plasma with the indicated SARS-CoV-2 variant. Dots indicate convalescent sera (grey), negative sera (black) and a serum from an immunocompromised individual with a lasting SARS-CoV-2 infection (green). (B) A549-AT cells were incubated with serially diluted (1:2) sera from vaccinated individuals together with the indicated SARS-CoV-2 variant. Dots indicate individual sera from BNT162b2 (blue), mRNA1273 (red) vaccinated individuals and SARS-CoV-2 negative sera (black). Additionally, a serum from a convalescent and BNT162b2-vaccinated individual (green) is shown. Statistical significance compared to FFM1 (B) and FFM7 (B.1) was calculated by two-tailed, paired student’s t-tests. Green dots (A+B) were excluded from significance testing. Mean values are depicted from two replicates. Asterisks indicate p-values as * (p < 0.05), ** (p ≤ 0.01), and *** (p ≤ 0.001).
Study significance
The study signifies that SARS-CoV-2 variants containing L452R spike mutation are more resistant to antibody-mediated neutralization than previously circulating variants with D614G spike mutation. Furthermore, compared to the co-circulating kappa variant, the delta variant has lower susceptibility to neutralization by antibodies induced by vaccination or infection.
Among currently available monoclonal antibodies, casirivimab is not effective against the kappa variant containing E484Q mutation. In contrast, the delta variant with T478K mutation exhibits resistance to imdevimab-mediated neutralization. Importantly, combination therapy with imdevimab and casirivimab offers effective neutralization of both kappa and delta variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Wilhelm A. 2021. Antibody mediated neutralization of authentic SARS-CoV-2 B.1.617 variants harboring L452R and T478K/E484Q, medRxiv, https://doi.org/10.1101/2021.08.09.21261704, https://www.medrxiv.org/content/10.1101/2021.08.09.21261704v2
- Peer reviewed and published scientific report.
Wilhelm, Alexander, Tuna Toptan, Christiane Pallas, Timo Wolf, Udo Goetsch, Rene Gottschalk, Maria J. G. T. Vehreschild, Sandra Ciesek, and Marek Widera. 2021. “Antibody-Mediated Neutralization of Authentic SARS-CoV-2 B.1.617 Variants Harboring L452R and T478K/E484Q.” Viruses 13 (9): 1693. https://doi.org/10.3390/v13091693. https://www.mdpi.com/1999-4915/13/9/1693.