The coronavirus disease 2019 (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The current COVID-19 pandemic has challenged the global health and economy, causing an unprecedented impact on the world in its wake.
Scientists have implemented several different diagnostic strategies to monitor the transmission of SARS-CoV-2 and inform public health measures. The current tests used to diagnose COVID-19 include those that detect the nucleic acids or proteins of SARS-CoV-2.
Both widespread testing and large-scale vaccination have successfully reduced the prevalence and severity of COVID-19 in some parts of the world. Despite these efforts, the emergence of SARS-CoV-2 variants of concern (VoC) such as the alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2) variants, have challenged the ability to contain SARS-CoV-2. Many VoCs have higher transmission rates and/or the capability to partially resist vaccines and therapeutics as compared to the original SARS-CoV-2 strain.
Whole-genome sequencing and SARS-CoV-2
VoCs may clearly pose a threat to the fight against the pandemic; therefore, genomic monitoring is essential to track the continuously mutating SARS-CoV-2. Whole-genome sequencing (WGS), of the approximately 30,000 base genome of SARS-CoV-2 is often used for genomic surveillance. WGS involves RNA extraction, molecular library preparation, and deep sequencing.
The main limitation associated with WGS is that it is resource-, cost-, and time-intensive. Furthermore, WGS is unevenly applied across countries, which has caused the number of genomes sequenced (more than 2 million) to be significantly smaller as compared to the global number of infections (~183 million), as of June 2021.
Therefore, it is crucial to increase the throughput and capacity for genomic surveillance of SARS-CoV-2 to track and prevent the spread of VoCs. A new study published on the medRxiv* preprint server discusses the development of DeepSARS, a high-throughput platform for simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2.
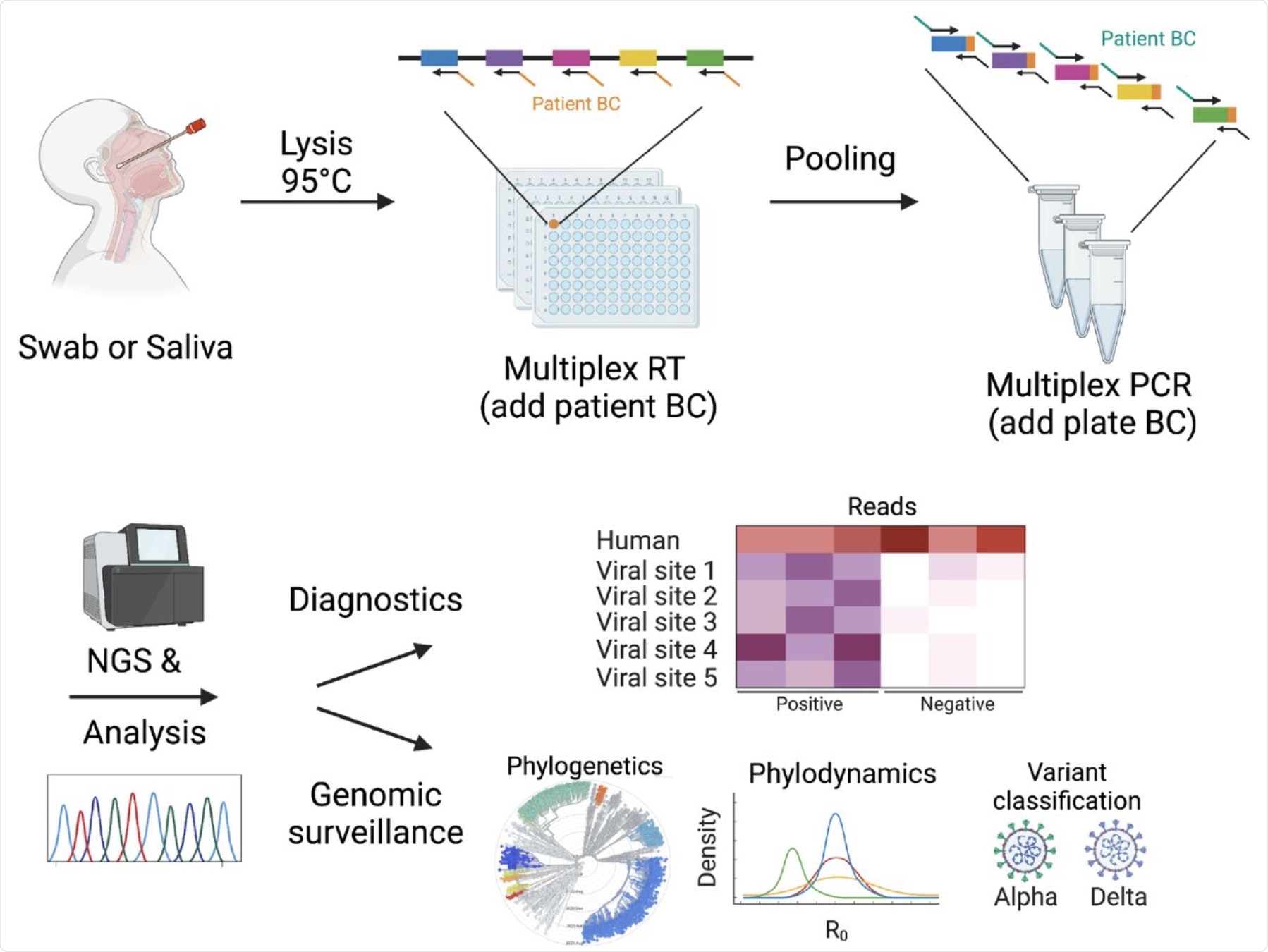 DeepSARS uses molecular barcodes (BCs) and multiplexed targeted deep sequencing (NGS) to enable simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2.
DeepSARS uses molecular barcodes (BCs) and multiplexed targeted deep sequencing (NGS) to enable simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2.
Study findings
In the current study, the researchers have developed DeepSARS, which is a quick and scalable approach that enables the simultaneous detection of VoCs, as well as the real-time monitoring of viral evolution.
DeepSARS is based on molecular barcoding, as well as targeted deep and computational phylogenetics. Therefore, this technology augments basic diagnostic testing capacity with genomic surveillance.
Herein, the scientists have shown that DeepSARS has excellent sensitivity and specificity to SARS-CoV-2 ribonucleic acid (RNA) material isolated from both nasopharyngeal swabs and saliva samples. Notably, DeepSARS appears to be sensitive to RNA extraction processing, freeze-thaw cycles, and storage conditions.
The stability of these samples while using DeepSARS is notable, as previous research has shown that storage conditions can significantly alter the composition of RNA samples. For example, one recent study found that human RNA (hRNA) was reduced; however, the detection of viral RNA was not impacted. DeepSARS generates a ratio of viral RNA:hRNA; therefore, a lower human-read may lead to high ratios which, in turn, could increase the false positive rate.
DeepSARS is still in its early stages, with future iterations of this technology expected to exhibit improved sensitivity and accuracy characteristics. Although the researchers here were able to investigate the relationship between the total number of human and viral reads, the accuracy of these readings can be greatly increased by making some modifications, such as incorporating site-specific information and deploying machine learning methods.
The current research develops a proof-of-concept that evaluates the performance of DeepSARS on a small subset of patient samples, relying on relatively fast but lower read count deep sequencing runs. As previously mentioned, DeepSARS is scalable and can be applied to hundreds to thousands of patients.
Some further technological advancements, like incorporating automated workflows and using multiple machines in parallel, could further enhance the testing capacity of DeepSARS. The theoretical throughput could be compared to other targeted deep sequencing-based diagnostics. Overall, the increased genomic and sequence space coverage of DeepSARs is expected to enhance genomic surveillance projects.
The use of DeepSARS resulted in comparable phylodynamic estimates relative to WGS. At the same time, it was observed that tree topologies diverged from those inferred using WGS. For DeepSARS to play a crucial role in shaping public health policies, it would need to accurately estimate the reproductive number (Re), as VoCs often have higher Re and transmission rates.
Conclusion
A major advantage associated with DeepSARS is its ability to rapidly adapt the protocol, as new primers can be easily introduced to cover emerging variants. Here, the scientists demonstrated three different parameters to which primers could be designed and implemented in DeepSARS. These parameters include mutational divergence from WGS, targeting specific VoCs, and targeting viral regions, such as the spike protein and its receptor-binding domain (RBD).
The design and validation of primers were completed before the emergence of the delta and gamma variants; however, the viral regions currently recovered by DeepSARS are capable of classifying such novel variants. This implies that DeepSARS could provide early warnings regarding the emergence of future variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Yermanos, A., Hong, K., Agrafiotis, A., et al. (2021) DeepSARS: simultaneous diagnostic detection and genomic surveillance of SARS-CoV-2. medRxiv. doi:10.1101/2021.08.16.21262126. https://www.medrxiv.org/content/10.1101/2021.08.16.21262126v1.
- Peer reviewed and published scientific report.
Yermanos, Alexander, Kai-Lin Hong, Andreas Agrafiotis, Jiami Han, Sarah Nadeau, Cecilia Valenzuela, Asli Azizoglu, et al. 2022. “DeepSARS: Simultaneous Diagnostic Detection and Genomic Surveillance of SARS-CoV-2.” BMC Genomics 23 (1). https://doi.org/10.1186/s12864-022-08403-0. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-022-08403-0.