Severe cases of coronavirus disease (COVID-19), which is caused by SARS-CoV-2, are associated with extreme inflammatory processes in the lung that can result in acute respiratory distress syndrome, lung or multi-organ failure – and even death.
Previous studies have shown that p38/MAPK pathway becomes activated during the infection with the aforementioned virus and that the inhibition of p38 can curb inflammatory cytokine expression and viral replication, implying that p38 inhibition may actually target multiple mechanisms related to SARS-CoV-2 pathogenesis.
However, even though the exact mechanisms by which the p38/MAPK pathway of our body is able to modulate inflammatory cytokine gene expression and RNA stability are described in-depth, it is still a mystery how p38/MAPK inhibition can reduce SARS-CoV-2 replication.
In addition, there are four different isoforms of p38 kinase (i.e., proteins similar to each other that carry out analogous roles within cells) and different downstream effector kinases, so we still do not know which kinases at which levels of the p38/MAPK pathway can impact the replication of SARS-CoV-2.
Consequently, this has piqued the interest of a group of scientists led by Dr. Christina A. Higgins and Dr. Jeffrey R. Johnson from the Icahn School of Medicine at Mount Sinai in New York (USA). They used a plethora of state-of-the-art methods to answer these pertinent questions.
A comprehensive research approach
In this study, the researchers have combined small interfering RNA (siRNA) screening, quantitative proteomics, as well as chemical and genetic perturbations in order to elucidate interactions between the p38/MAPK pathway and SARS-CoV-2 in infected cells.
More specifically, four isoforms of p38 kinase (p38α, p38β, p38γ, p38δ) were appraised in-depth, and putative p38ß substrates have been identified in an unbiased manner, with substantial relevance beyond SARS-CoV-2 biology. In addition, Gene ontology enrichment and kinase activity analyses have also been pursued.
In short, their comprehensive research approach covered a wide array of classical virology techniques (such as plaque assays for counting infectious particles directly) and relatively modern approaches such as genetic sequencing and mass spectrometry.
A key player in SARS-CoV-2 replication
This study has found that several components of the p38/MAPK pathway can positively and negatively impact SARS-CoV-2 infection. Overall, the activity of this pathway is substantially increasing during the infection with SARS-CoV-2 in human lung cells.
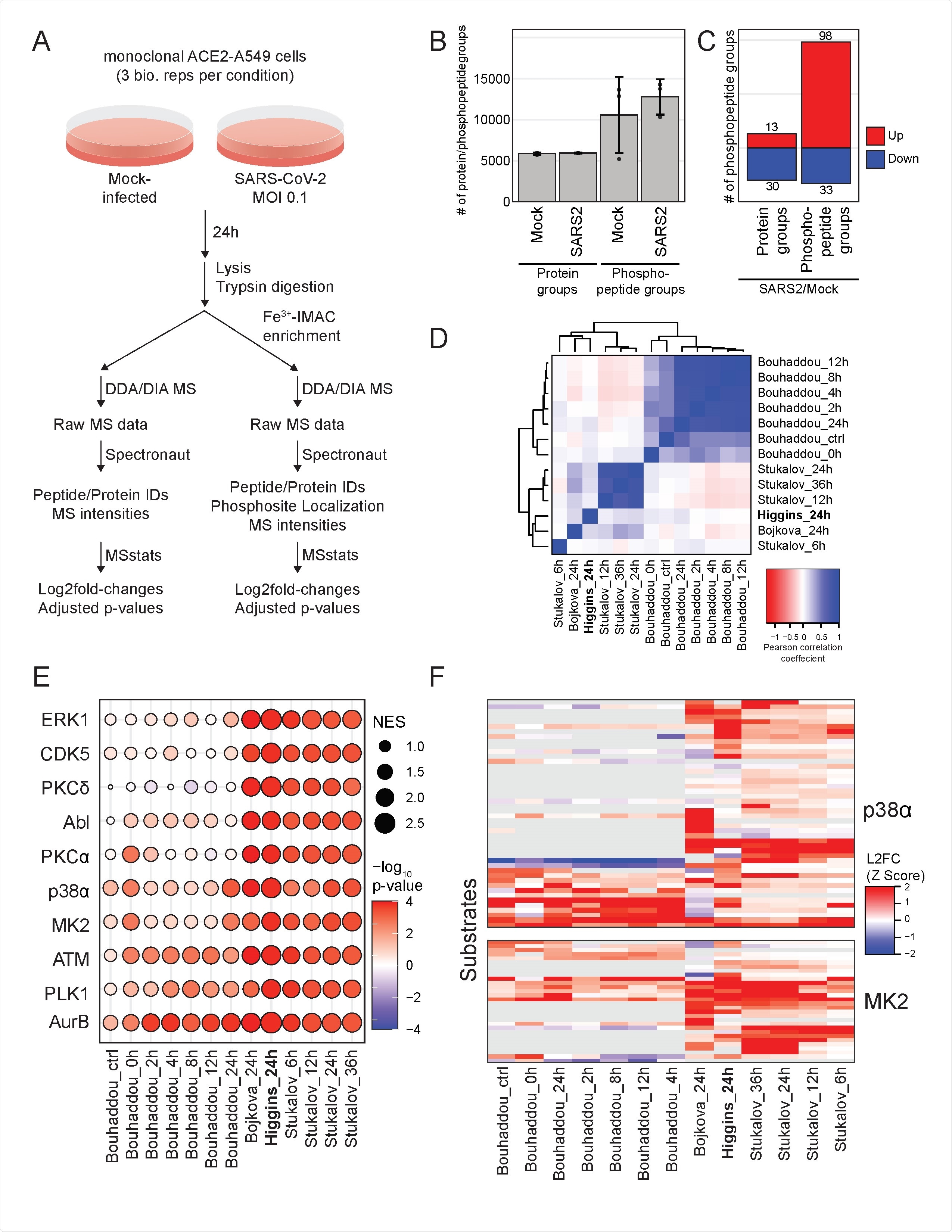
Meta-analysis of SARS-CoV-2 proteomics studies reveals pathways consistently regulated across species and cell types. A) Schematic of experimental design. B) Average of protein groups and of phosphorylation site groups quantified in each condition. Error bars are standard deviation. C) Number of differentially expressed protein groups and phosphorylation site groups for SARS-CoV-2 infected cells fold over mock-infected cells D) Pairwise Pearson coefficients of protein abundance log2fold-change profiles from this study and published studies indicated. E) Kinase activity analysis based on log2fold-change profiles from this study and published studies indicated. The absolute value of the normalized enrichment score (NES) is indicated by node sizes and the -Log10(p-value) is indicated by the color scale. Decreases in kinase activity are indicated by negative -LogP values. F) Z-score-transformed log2fold-change profiles of p38/MAPK kinases p38α and MK2 in this study and published studies indicated. Each row represents a protein substrate of the respective kinase with the maximum log2fold-change of all phosphorylation sites each substrate indicated by the color scale.
From kinase isoforms that have been tested, p38ß was found to be an indispensable host factor for SARS-CoV-2 replication in human lung epithelial cells, but also for viral protein translation without innate immune sensing. The researchers have also found that p38 inhibition can reduce viral protein levels but does not impact levels of viral messenger RNA (mRNA).
Finally, p38 inhibition was shown to diminish phosphorylation levels at manifold sites on the SARS-CoV-2 nucleocapsid (or N) protein, which is a multifunctional structure that binds to the viral RNA genome and packs it into a long helical structure.
Implications for treating COVID-19
These findings not only improve our understanding of fundamental SARS-CoV-2 biology, but can also be used to identify novel drug targets for treating COVID-19 and to pave the way for the identification of kinase substrates that can be widely applied in different research areas.
“These data, in addition to other p38ß-focused research, emphasize the need for p38ß-specific inhibitors and reagents to help better characterize this isoform”, say study authors in this bioRxiv preprint paper.
“p38ß does not appear to be essential, and it has a distinctly smaller active site than p38α that may allow for the development of specific inhibitors, thus we believe p38ß may make an attractive target”, they add.
In any case, further studies are needed to solve the puzzle of p38ß promoting viral protein translation, as well as to appraise its role in the replication of other coronaviruses, but also other families of potentially emerging viral agents.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.