The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease (COVID-19) pandemic, has made it imperative to have rapid, sensitive, specific, and portable testing which is deployable immediately in a point-of-care context for instant detection of the virus.
 Study: CRISPR-Cas based virus detection: Recent advances and perspectives. Image Credit: nobeastsofierce/ Shutterstock
Study: CRISPR-Cas based virus detection: Recent advances and perspectives. Image Credit: nobeastsofierce/ Shutterstock
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated proteins (Cas) – based systems of molecular diagnosis of viral proteins have been researched widely. The CRISPR-Cas protein constitutes a nucleic- acid-based adaptive immune system in bacteria and archaea, which used RNA-guided nucleases to cleave invading nucleic acids. Scientists have utilized this phenomenon, and the mechanism is used to detect viral genetic material.
CRISPR-Cas systems, especially CRISPR-Cas12a and CRISPR-Cas13a, are distinctively known for their sensitivity, specificity, high base resolution, and programmability for recognizing specific sequences of nucleic acids. They have been used to identify viruses like Zika, Ebola, Influenza A, and Human Papilloma Virus.
Researchers from China recently reviewed the latest advancements, applications, and prospects of virus detection using CRISPR-Cas systems, especially CRISPR-Cas12a/Cas13a, in a review in Biosensors and Bioelectronics.
What is CRISPR and how does it help detect viral infections?
The Cas constitutes a nucleic acid-based gene recognition machinery, programmed to recognize and cleave specific parts of the viral genetic material or RNA and release various signals in parallel for a machine to read and analyze the results.
The most popular CRISPR Cas protein systems are the type II CRISPR systems with Cas9, type V systems with Cas12a (also known as cpf1), and type VI systems with Cas13a. The main guide RNA (sgRNA) recognizes a short specific sequence termed PAM (Proto-spacer Adjacent Motif), which helps the Cas9 recognize and cleave the specific double-stranded DNA (dsDNA), breaking the DNA structure with the help of specific proteins present in the Cas9.
The main mechanism of CRISPR Cas-based viral genome cleaving remains the same for both CRISPR Cas12a and CRISPR Cas13a systems. However, both types of machinery can detect two different types of viral genetic material,
- the CRISPR Cas12a detects single-stranded (ssDNA) or dsDNA, which, in the case of RNA viruses, needs to be reverse-transcribed from RNA
- the CRISPR Cas13a system can readily recognize single-stranded RNA (ssRNA), which needs no reverse transcription.
- Another advantage of the CRISPR Cas13a system is that it needs a relatively simple specific sequence, known as protospacer flanking site (PFS), in comparison to CRISPR Cas12a,
What is the significance and importance of CRISPR in viral detection?
The CRISPR Cas13a system was utilized to make a Specific High-Sensitivity Enzymatic Reporter Unlocking (SHERLOCK) technique to detect viral genetic material in patient samples. Another diagnostic system, known as DNA Endonuclease Targeted CRISPR Trans Reporter (DETECTR) was made, using the CRISPR-Cas12a system. Both SHERLOCK and DETECTR systems have been used in several ways with different signal analysis systems to detect different forms of viruses (plant, animal, and human viruses).
Some prime examples include,
- Detection of Zika Virus, Dengue Virus, variants of Human Papilloma Virus (HPV) and even Human Immunodeficiency Virus (HIV), Ebola Virus, Lassa Virus, Influenza A virus, African Swine Fever Virus, Canine Parvovirus, White Spot Syndrome Virus, Tobacco Curly Shoot Virus, and more recently the SARS-CoV-2 – all of which employed a fluorophore-quencher complex along with the CRISPR Cas12a and 13a proteins, which released fluorescent signals to be read by either microplate readers, smartphone sensors, microfluidic chips, 3D printing instruments or LED lights
- Detection of African Swine Fever Virus, Apple Stem Pitting and Grooving virus, Rapevine red blotch virus, using colorimetric signals using gold nanoparticles which can be detected by the naked eye, as well as spectrophotometers (by measuring the absorption spectra)
- Human papillomavirus and Parvovirus variants have also been detected using signal transduction pathways and generation of electrochemical signals on detection of the specific protein
- Viruses have even been identified using Lateral Flow Assays that involve antigen-antibody interaction or that between target DNA sequence and the probe DNA, using conjugated gold, carbon, or colored latex nanoparticles within the conjugate pad
- Owing to the high sensitivity and specificity of the methods, they have been modified to specifically detect mutants of the viruses by altering very specific sequences in the probe DNA / RNA material; this becomes crucial in testing for mutating variants of novel viruses, such as the SARS Coronavirus
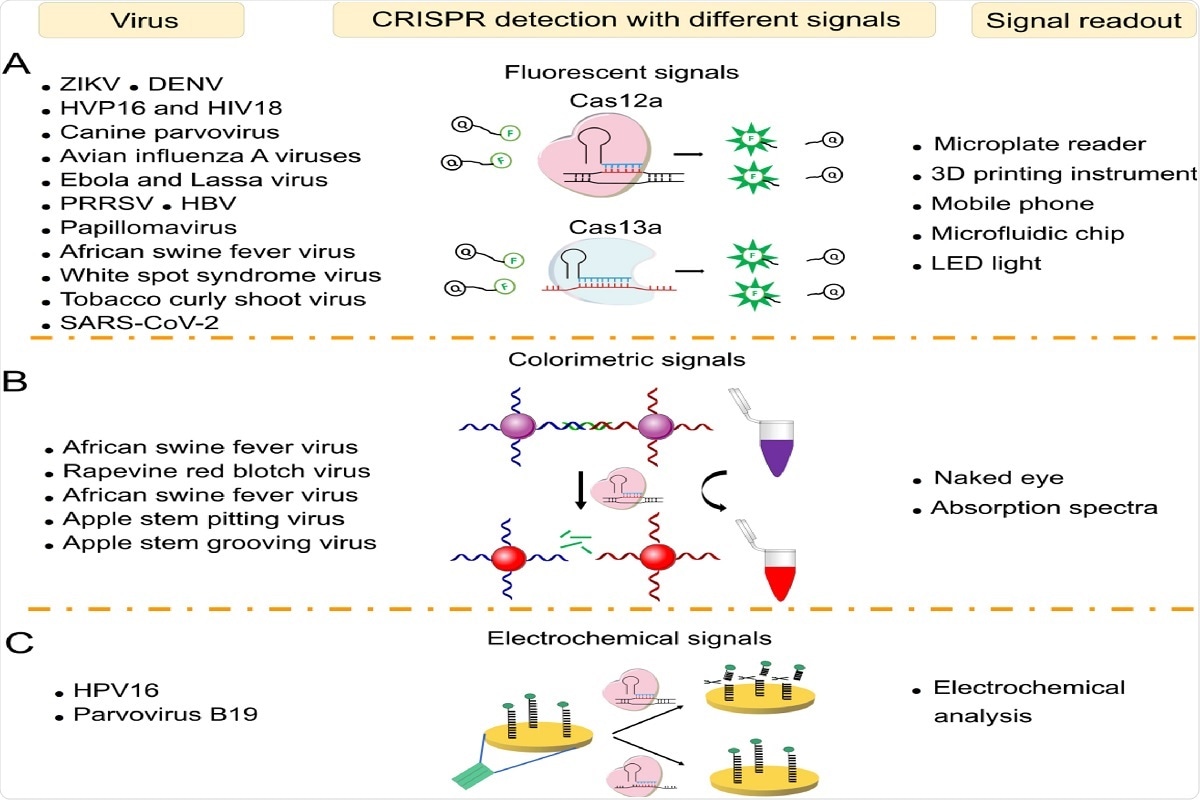 Fig. 3. Virus sensing with different signal readouts. (A) Virus sensing based on CRISPR-Cas12a/Cas13a via fluorescent signals. (B) Virus sensing via colorimetric signals. (C) Virus sensing via electrochemical signals.
Fig. 3. Virus sensing with different signal readouts. (A) Virus sensing based on CRISPR-Cas12a/Cas13a via fluorescent signals. (B) Virus sensing via colorimetric signals. (C) Virus sensing via electrochemical signals.
What are the challenges and perspectives of CRISPR in the future of viral detection?
It is now possible to detect viral genetic material owing to the high resolution, sensitivity, and specificity of the CRISPR Cas 12a and 13a systems. From a resolution perspective, they can be used in detecting genetic material at femtomolar (fM) (without amplification) or attomolar (aM) (with amplification) levels. Even mutations at the level of one single nucleotide can be detected using this revolutionary diagnostic tool.
There have been challenges in the form of lack of standardization methods, contamination-free sample preparation and pre-treatment, obtaining an accurate quantified analysis of viral load from samples, and even getting desired lengths of target sequences for deploying in the diagnostic tests.
CRISPR Cas-systems hold immense potential not only in viral genome detection but also in gene modification and editing. It can revolutionize biosensing and molecular diagnostics for the best outcomes in the future.